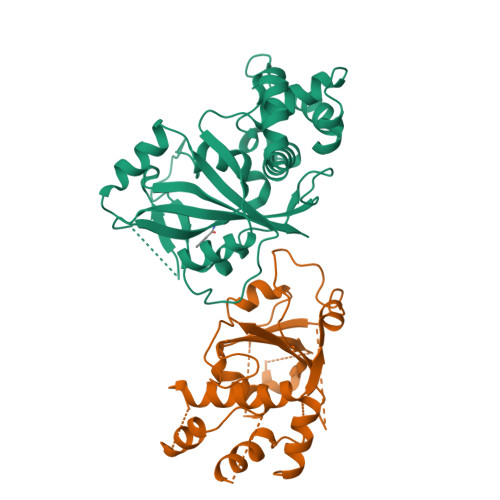
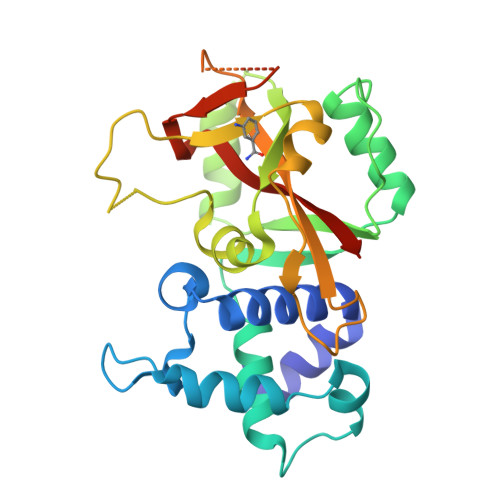
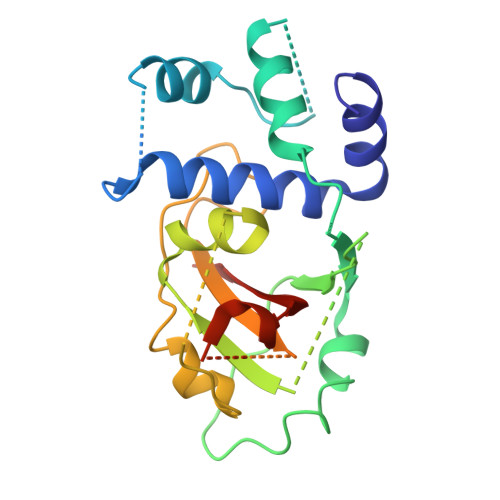
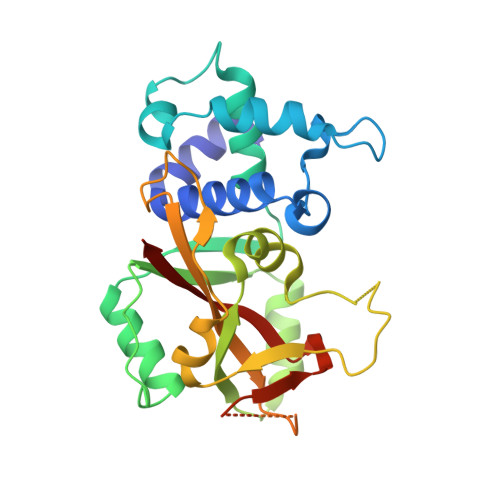
Crystal Structure of Human ADP-ribose Transferase ARTD15/PARP16 Reveals a Novel Putative Regulatory Domain.
Karlberg, T., Thorsell, A.G., Kallas, A., Schuler, H.(2012) J Biological Chem 287: 24077-24081
- PubMed: 22661712
- DOI: https://doi.org/10.1074/jbc.M112.379289
- Primary Citation of Related Structures:
4F0D - PubMed Abstract:
ADP-ribosylation is involved in the regulation of DNA repair, transcription, and other processes. The 18 human ADP-ribose transferases with diphtheria toxin homology include ARTD1/PARP1, a cancer drug target. Knowledge of other family members may guide therapeutics development and help evaluate potential drug side effects. Here, we present the crystal structure of human ARTD15/PARP16, a previously uncharacterized enzyme. ARTD15 features an α-helical domain that packs against its transferase domain without making direct contact with the NAD(+)-binding crevice or the donor loop. Thus, this novel domain does not resemble the regulatory domain of ARTD1. ARTD15 displays auto-mono(ADP-ribosylation) activity and is affected by canonical poly(ADP-ribose) polymerase inhibitors. These results add to a framework that will facilitate research on a medically important family of enzymes.
Organizational Affiliation:
Structural Genomics Consortium, Karolinska Institutet, 17177 Stockholm, Sweden.