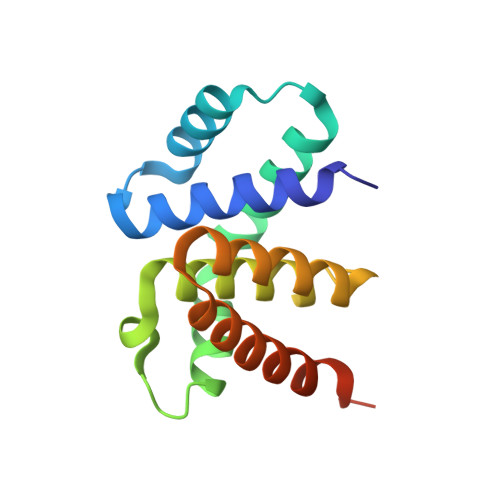
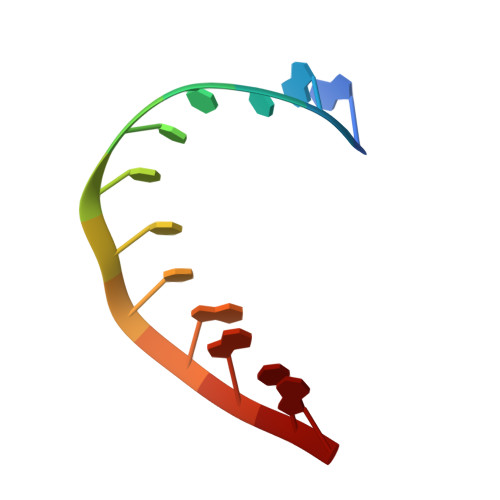
Crystal structure of a plectonemic RNA supercoil.
Stagno, J.R., Ma, B., Li, J., Altieri, A.S., Byrd, R.A., Ji, X.(2012) Nat Commun 3: 901-901
- PubMed: 22692544
- DOI: https://doi.org/10.1038/ncomms1903
- Primary Citation of Related Structures:
4EYA - PubMed Abstract:
Genome packaging is an essential housekeeping process in virtually all organisms for proper storage and maintenance of genetic information. Although the extent and mechanisms of packaging vary, the process involves the formation of nucleic-acid superstructures. Crystal structures of DNA coiled coils indicate that their geometries can vary according to sequence and/or the presence of stabilizers such as proteins or small molecules. However, such superstructures have not been revealed for RNA. Here we report the crystal structure of an RNA supercoil, which displays one level higher molecular organization than previously reported structures of DNA coiled coils. In the presence of an RNA-binding protein, two interlocking RNA coiled coils of double-stranded RNA, a 'coil of coiled coils', form a plectonemic supercoil. Molecular dynamics simulations suggest that protein-RNA interaction is required for the stability of the supercoiled RNA. This study provides structural insight into higher order packaging mechanisms of nucleic acids.
Organizational Affiliation:
Macromolecular Crystallography Laboratory, National Cancer Institute, Frederick, Maryland 21702, USA.