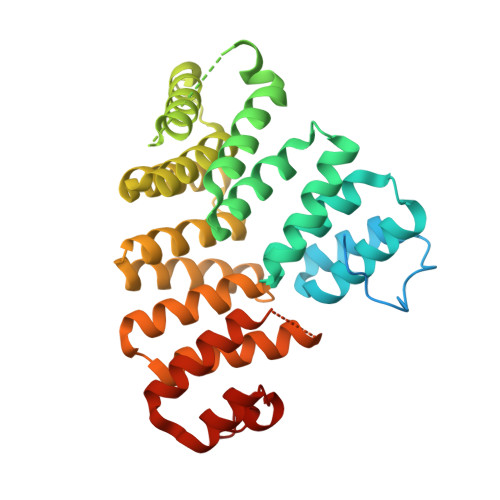
Structure and stoichiometry of an accessory subunit TRIP8b interaction with hyperpolarization-activated cyclic nucleotide-gated channels.
Bankston, J.R., Camp, S.S., Dimaio, F., Lewis, A.S., Chetkovich, D.M., Zagotta, W.N.(2012) Proc Natl Acad Sci U S A 109: 7899-7904
- PubMed: 22550182
- DOI: https://doi.org/10.1073/pnas.1201997109
- Primary Citation of Related Structures:
4EQF - PubMed Abstract:
Ion channels operate in intact tissues as part of large macromolecular complexes that can include cytoskeletal proteins, scaffolding proteins, signaling molecules, and a litany of other molecules. The proteins that make up these complexes can influence the trafficking, localization, and biophysical properties of the channel. TRIP8b (tetratricopetide repeat-containing Rab8b-interacting protein) is a recently discovered accessory subunit of hyperpolarization-activated cyclic nucleotide-gated (HCN) channels that contributes to the substantial dendritic localization of HCN channels in many types of neurons. TRIP8b interacts with the carboxyl-terminal region of HCN channels and regulates their cell-surface expression level and cyclic nucleotide dependence. Here we examine the molecular determinants of TRIP8b binding to HCN2 channels. Using a single-molecule fluorescence bleaching method, we found that TRIP8b and HCN2 form an obligate 4:4 complex in intact channels. Fluorescence-detection size-exclusion chromatography and fluorescence anisotropy allowed us to confirm that two different domains in the carboxyl-terminal portion of TRIP8b--the tetratricopepide repeat region and the TRIP8b conserved region--interact with two different regions of the HCN carboxyl-terminal region: the carboxyl-terminal three amino acids (SNL) and the cyclic nucleotide-binding domain, respectively. And finally, using X-ray crystallography, we determined the atomic structure of the tetratricopepide region of TRIP8b in complex with a peptide of the carboxy-terminus of HCN2. Together, these experiments begin to uncover the mechanism for TRIP8b binding and regulation of HCN channels.
Organizational Affiliation:
Department of Physiology and Biophysics, University of Washington School of Medicine, Seattle, WA 98195, USA.