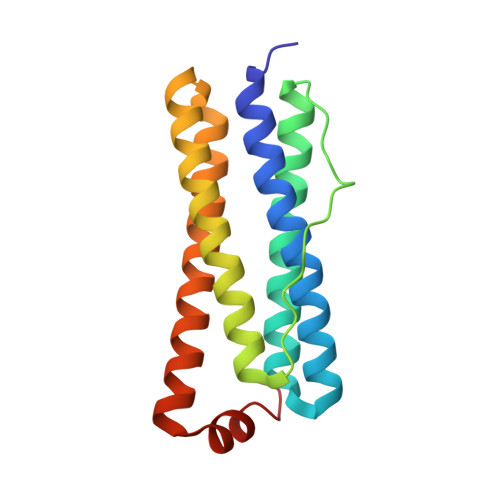
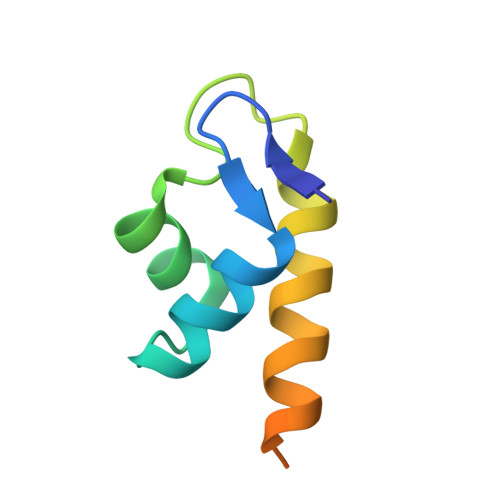
The Structure of the BfrB-Bfd Complex Reveals Protein-Protein Interactions Enabling Iron Release from Bacterioferritin.
Yao, H., Wang, Y., Lovell, S., Kumar, R., Ruvinsky, A.M., Battaile, K.P., Vakser, I.A., Rivera, M.(2012) J Am Chem Soc 134: 13470-13481
- PubMed: 22812654
- DOI: https://doi.org/10.1021/ja305180n
- Primary Citation of Related Structures:
4E6K - PubMed Abstract:
Ferritin-like molecules are unique to cellular iron homeostasis because they can store iron at concentrations much higher than those dictated by the solubility of Fe(3+). Very little is known about the protein interactions that deliver iron for storage or promote the mobilization of stored iron from ferritin-like molecules. Here, we report the X-ray crystal structure of Pseudomonas aeruginosa bacterioferritin (Pa-BfrB) in complex with bacterioferritin-associated ferredoxin (Pa-Bfd) at 2.0 Å resolution. As the first example of a ferritin-like molecule in complex with a cognate partner, the structure provides unprecedented insight into the complementary interface that enables the [2Fe-2S] cluster of Pa-Bfd to promote heme-mediated electron transfer through the BfrB protein dielectric (~18 Å), a process that is necessary to reduce the core ferric mineral and facilitate mobilization of Fe(2+). The Pa-BfrB-Bfd complex also revealed the first structure of a Bfd, thus providing a first view to what appears to be a versatile metal binding domain ubiquitous to the large Fer2_BFD family of proteins and enzymes with diverse functions. Residues at the Pa-BfrB-Bfd interface are highly conserved in Bfr and Bfd sequences from a number of pathogenic bacteria, suggesting that the specific recognition between Pa-BfrB and Pa-Bfd is of widespread significance to the understanding of bacterial iron homeostasis.
Organizational Affiliation:
Department of Chemistry, University of Kansas, Multidisciplinary Research Building, 2030 Becker Drive, Lawrence, Kansas 66047, USA.