Crystal Structure of endoribonuclease Cas6e from Escherichia coli
Wei, J., Huang, G., Wang, Y., Gong, W.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Uncharacterized protein ygcH | 218 | Escherichia coli K-12 | Mutation(s): 0 Gene Names: ygcH EC: 3.1 | 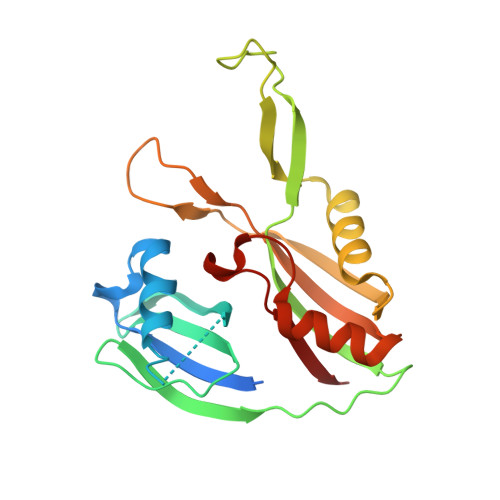 | |
UniProt | |||||
Find proteins for Q46897 (Escherichia coli (strain K12)) Explore Q46897 Go to UniProtKB: Q46897 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q46897 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GOL Query on GOL | B [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 59.9 | α = 90 |
| b = 59.9 | β = 90 |
| c = 128.66 | γ = 90 |
| Software Name | Purpose |
|---|---|
| ADSC | data collection |
| CNS | refinement |
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| CNS | phasing |