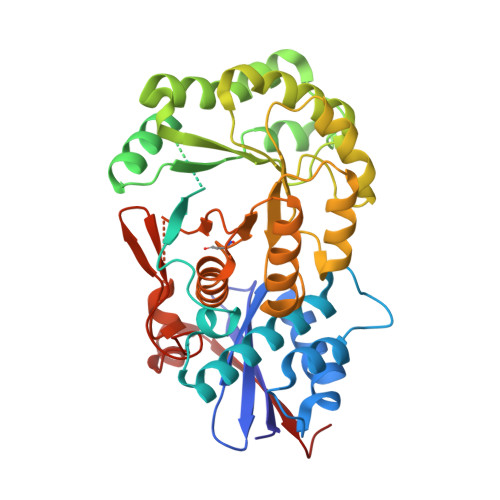
Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue
Vetting, M.W., Bouvier, J.T., Toro, R., Bhosle, R., Al Obaidi, N.F., Morisco, L.L., Wasserman, S.R., Sojitra, S., Imker, H.J., Gerlt, J.A., Almo, S.C., Enzyme Function Initiative (EFI)To be published.