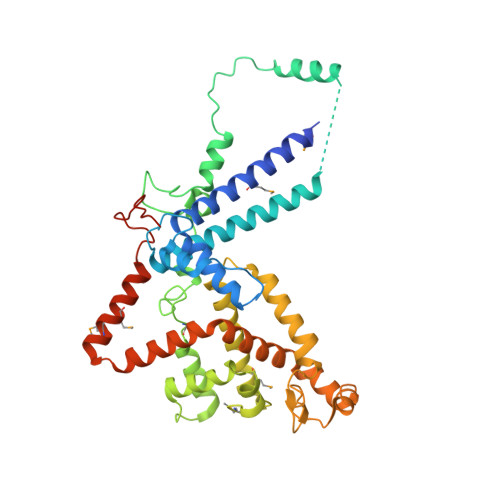
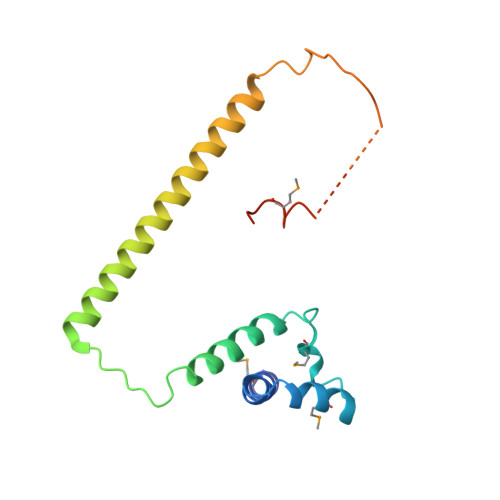
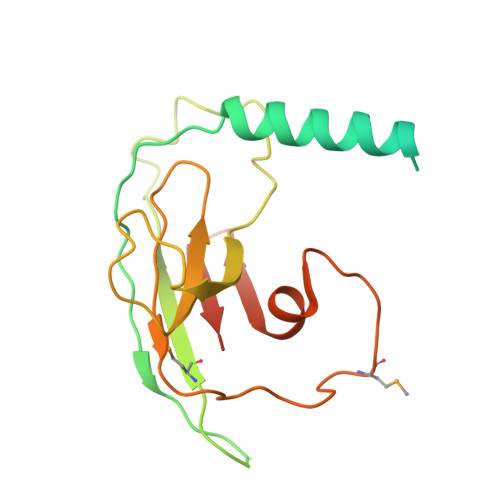
Structure and assembly of the SF3a splicing factor complex of U2 snRNP
Lin, P.C., Xu, R.M.(2012) EMBO J 31: 1579-1590
- PubMed: 22314233
- DOI: https://doi.org/10.1038/emboj.2012.7
- Primary Citation of Related Structures:
4DGW - PubMed Abstract:
SF3a is an evolutionarily conserved heterotrimeric complex essential for pre-mRNA splicing. It functions in spliceosome assembly within the mature U2 snRNP (small nuclear ribonucleoprotein particle), and its displacement from the spliceosome initiates the first step of the splicing reaction. We have identified a core domain of the yeast SF3a complex required for complex assembly and determined its crystal structure. The structure shows a bifurcated assembly of three subunits, Prp9, Prp11 and Prp21, with Prp9 interacting with Prp21 via a bidentate-binding mode, and Prp21 wrapping around Prp11. Structure-guided biochemical analysis also shows that Prp9 harbours a major binding site for stem-loop IIa of U2 snRNA. These findings provide mechanistic insights into the assembly of U2 snRNP.
Organizational Affiliation:
Structural Biology Program, The Kimmel Center for Biology and Medicine at the Skirball Institute of Biomolecular Medicine, and the Department of Pharmacology, New York University School of Medicine, New York, NY, USA.