Structure, Activity and Stereoselectivity of Nadph-Dependent Oxidoreductases Catalysing the S-Selective Reduction of the Imine Substrate 2-Methylpyrroline.
Man, H., Wells, E., Hussain, S., Leipold, F., Hart, S., Turkenburg, J.P., Turner, N.J., Grogan, G.(2015) Chembiochem 16: 1052
- PubMed: 25809902
- DOI: https://doi.org/10.1002/cbic.201402625
- Primary Citation of Related Structures:
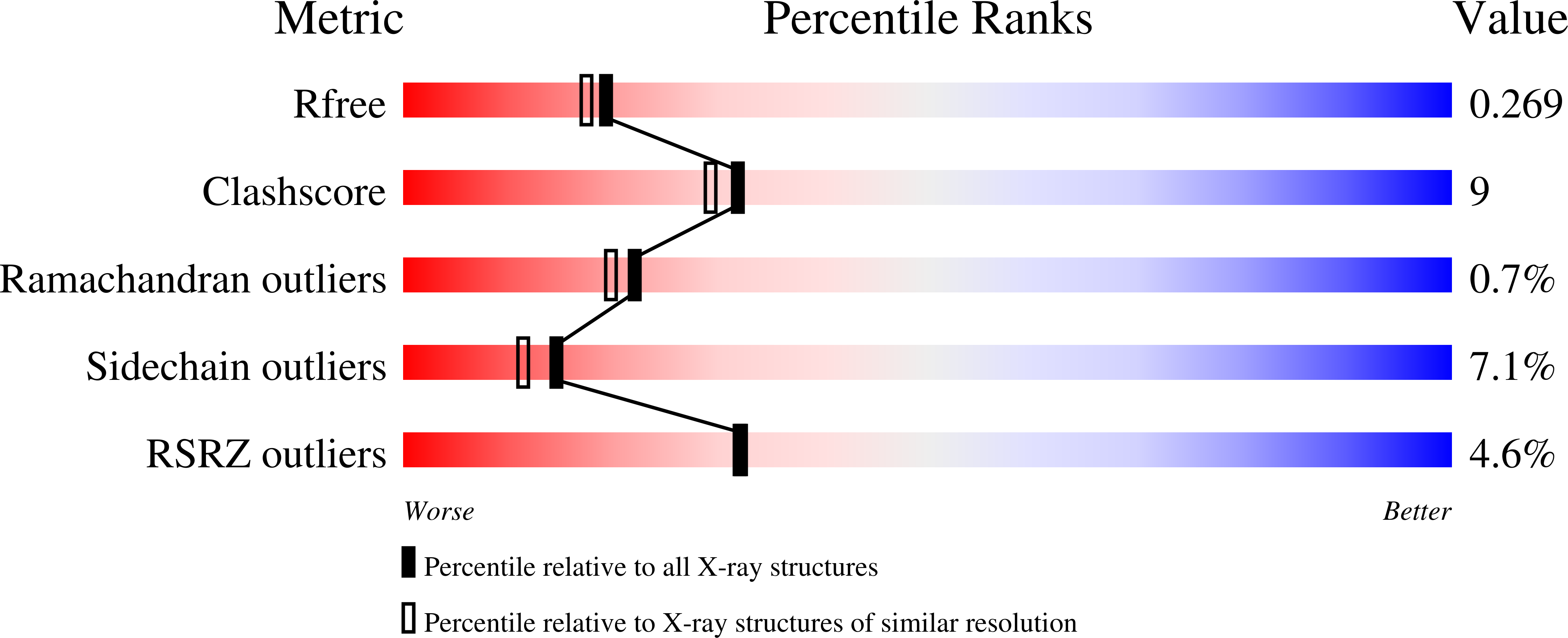
4D3D, 4D3F, 4D3S - PubMed Abstract:
Oxidoreductases from Streptomyces sp. GF3546 [3546-IRED], Bacillus cereus BAG3X2 (BcIRED) and Nocardiopsis halophila (NhIRED) each reduce prochiral 2-methylpyrroline (2MPN) to (S)-2-methylpyrrolidine with >95 % ee and also a number of other imine substrates with good selectivity. Structures of BcIRED and NhIRED have helped to identify conserved active site residues within this subgroup of imine reductases that have S selectivity towards 2MPN, including a tyrosine residue that has a possible role in catalysis and superimposes with an aspartate in related enzymes that display R selectivity towards the same substrate. Mutation of this tyrosine residue-Tyr169-in 3546-IRED to Phe resulted in a mutant of negligible activity. The data together provide structural evidence for the location and significance of the Tyr residue in this group of imine reductases, and permit a comparison of the active sites of enzymes that reduce 2MPN with either R or S selectivity.
Organizational Affiliation:
Department of Chemistry, University of York, Heslington, York, YO10 5DD (UK).