Structural and Functional Insights Into the Mode of Action of a Universally Conserved Obg Gtpase.
Feng, B., Mandava, C.S., Guo, Q., Wang, J., Cao, W., Li, N., Zhang, Y., Zhang, Y., Wang, Z., Wu, J., Sanyal, S., Lei, J., Gao, N.(2014) PLoS Biol 12: 1866
- PubMed: 24844575
- DOI: https://doi.org/10.1371/journal.pbio.1001866
- Primary Citation of Related Structures:
4CSU - PubMed Abstract:
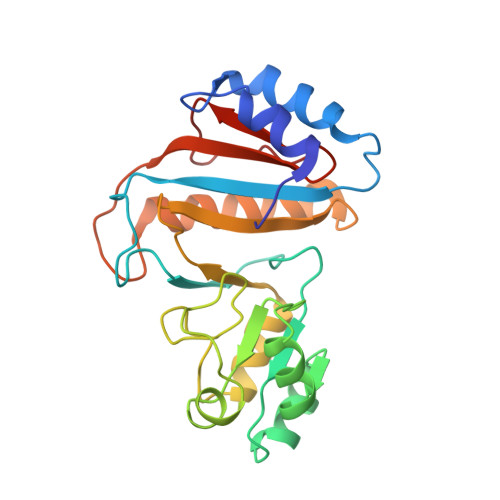
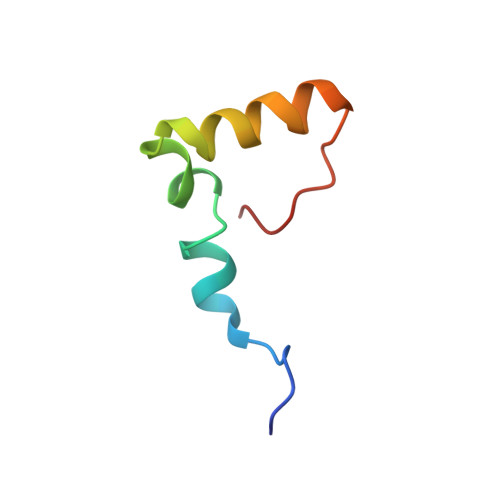
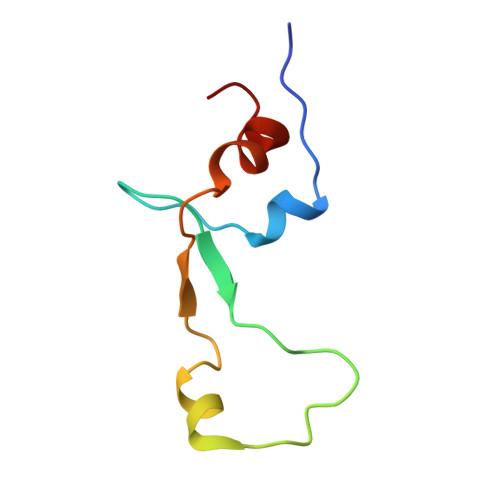
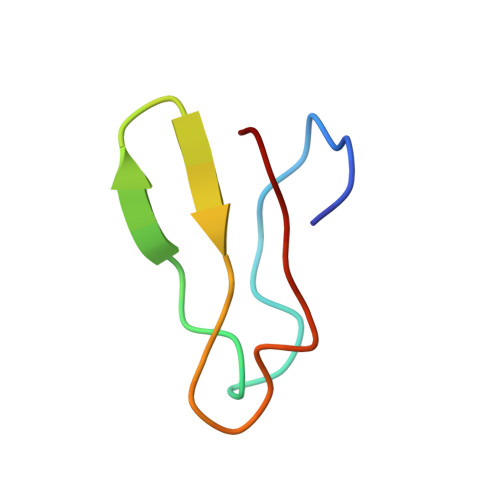
Obg proteins are a family of P-loop GTPases, conserved from bacteria to human. The Obg protein in Escherichia coli (ObgE) has been implicated in many diverse cellular functions, with proposed molecular roles in two global processes, ribosome assembly and stringent response. Here, using pre-steady state fast kinetics we demonstrate that ObgE is an anti-association factor, which prevents ribosomal subunit association and downstream steps in translation by binding to the 50S subunit. ObgE is a ribosome dependent GTPase; however, upon binding to guanosine tetraphosphate (ppGpp), the global regulator of stringent response, ObgE exhibits an enhanced interaction with the 50S subunit, resulting in increased equilibrium dissociation of the 70S ribosome into subunits. Furthermore, our cryo-electron microscopy (cryo-EM) structure of the 50S·ObgE·GMPPNP complex indicates that the evolutionarily conserved N-terminal domain (NTD) of ObgE is a tRNA structural mimic, with specific interactions with peptidyl-transferase center, displaying a marked resemblance to Class I release factors. These structural data might define ObgE as a specialized translation factor related to stress responses, and provide a framework towards future elucidation of functional interplay between ObgE and ribosome-associated (p)ppGpp regulators. Together with published data, our results suggest that ObgE might act as a checkpoint in final stages of the 50S subunit assembly under normal growth conditions. And more importantly, ObgE, as a (p)ppGpp effector, might also have a regulatory role in the production of the 50S subunit and its participation in translation under certain stressed conditions. Thus, our findings might have uncovered an under-recognized mechanism of translation control by environmental cues.
Organizational Affiliation:
Ministry of Education Key Laboratory of Protein Sciences, Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing, China.