Structure-Guided Simulations Illuminate the Mechanism of ATP Transport Through Vdac1.
Choudhary, O.P., Paz, A., Adelman, J.L., Colletier, J., Abramson, J., Grabe, M.(2014) Nat Struct Mol Biol 21: 626
- PubMed: 24908397
- DOI: https://doi.org/10.1038/nsmb.2841
- Primary Citation of Related Structures:
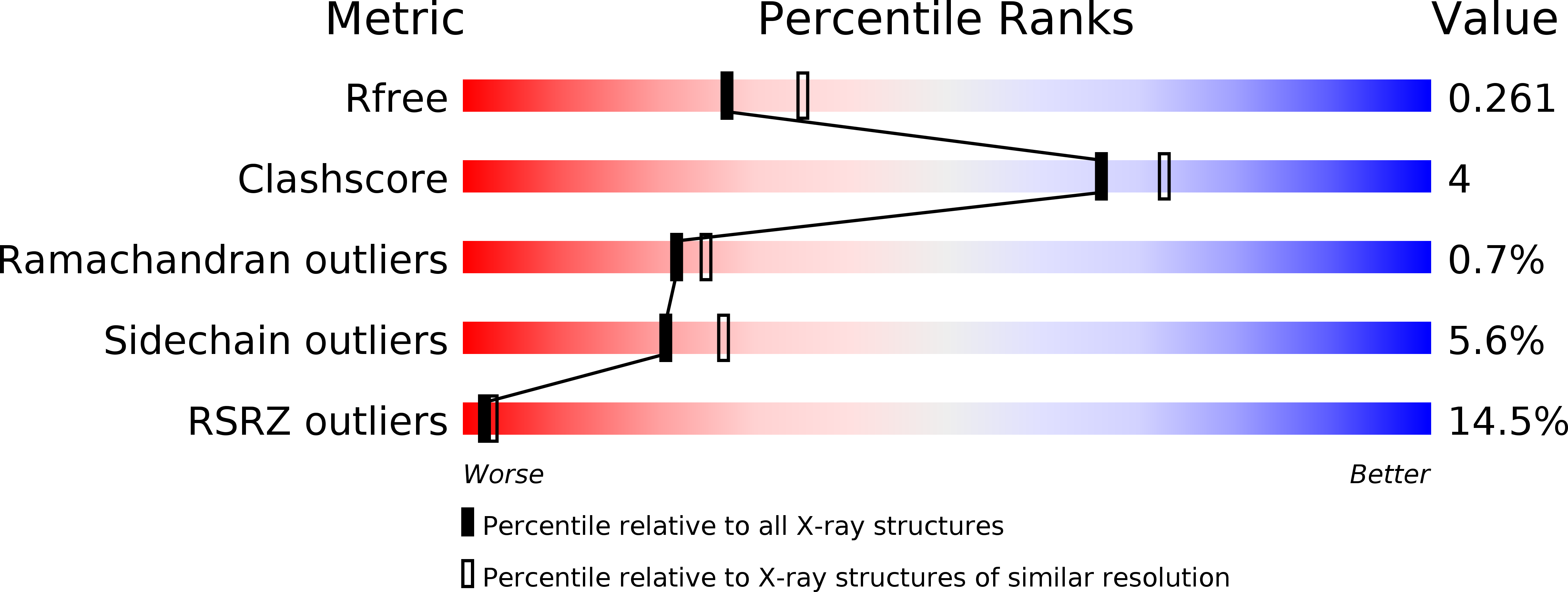
4C69 - PubMed Abstract:
The voltage-dependent anion channel (VDAC) mediates the flow of metabolites and ions across the outer mitochondrial membrane of all eukaryotic cells. The open channel passes millions of ATP molecules per second, whereas the closed state exhibits no detectable ATP flux. High-resolution structures of VDAC1 revealed a 19-stranded β-barrel with an α-helix partially occupying the central pore. To understand ATP permeation through VDAC, we solved the crystal structure of mouse VDAC1 (mVDAC1) in the presence of ATP, revealing a low-affinity binding site. Guided by these coordinates, we initiated hundreds of molecular dynamics simulations to construct a Markov state model of ATP permeation. These simulations indicate that ATP flows through VDAC through multiple pathways, in agreement with our structural data and experimentally determined physiological rates.
Organizational Affiliation:
1] Joint Carnegie Mellon University-University of Pittsburgh PhD Program in Computational Biology, Pittsburgh, Pennsylvania, USA. [2].