High-Resolution Structure and Double Electron-Electron Resonance of the Zebrafish Voltage Dependent Anion Channel 2 Reveal an Oligomeric Population.
Schredelseker, J., Paz, A., Lopez, C.J., Altenbach, C., Leung, C.S., Drexler, M.K., Chen, J., Hubbell, W.L., Abramson, J.(2014) J Biol Chem 289: 12566
- PubMed: 24627492
- DOI: https://doi.org/10.1074/jbc.M113.497438
- Primary Citation of Related Structures:
4BUM - PubMed Abstract:
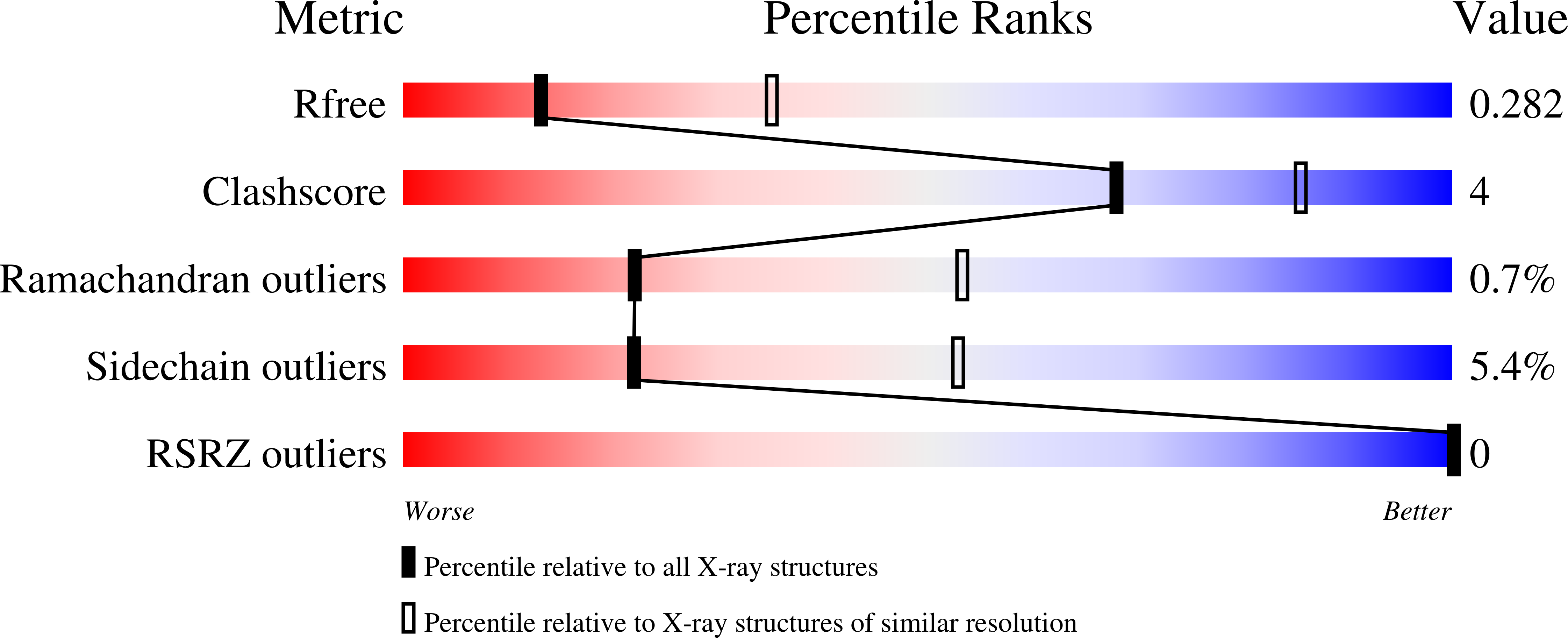
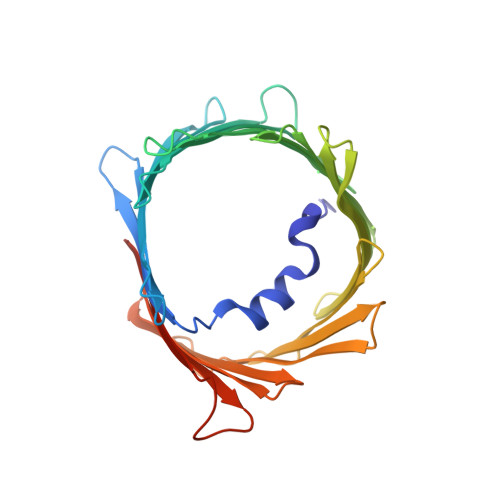
In recent years, there has been a vast increase in structural and functional understanding of VDAC1, but VDAC2 and -3 have been understudied despite having many unique phenotypes. One reason for the paucity of structural and biochemical characterization of the VDAC2 and -3 isoforms stems from the inability of obtaining purified, functional protein. Here we demonstrate the expression, isolation, and basic characterization of zebrafish VDAC2 (zfVDAC2). Further, we resolved the structure of zfVDAC2 at 2.8 Å resolution, revealing a crystallographic dimer. The dimer orientation was confirmed in solution by double electron-electron resonance spectroscopy and by cross-linking experiments disclosing a dimer population of ∼20% in lauryldimethine amine oxide detergent micelles, whereas in lipidic bicelles a higher population of dimeric and higher order oligomers species were observed. The present study allows for a more accurate structural comparison between VDAC2 and its better-studied counterpart VDAC1.
Organizational Affiliation:
From the Department of Molecular, Cell and Developmental Biology.