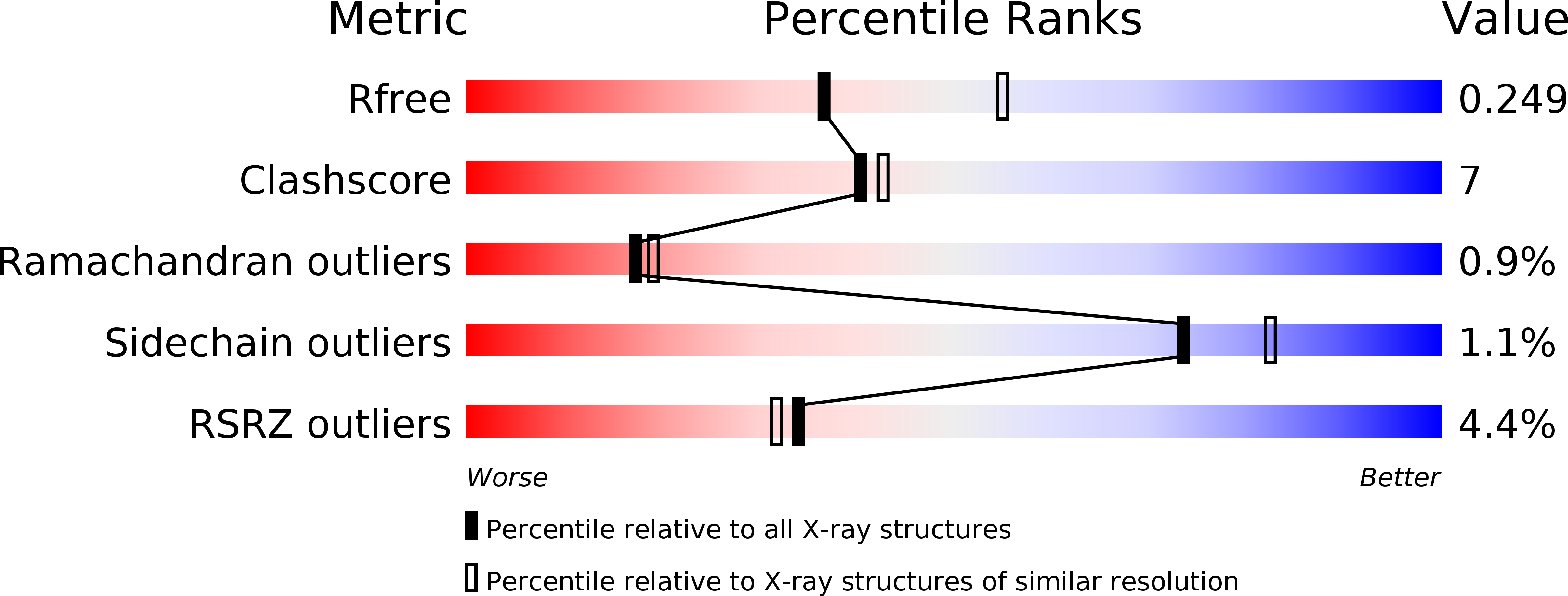
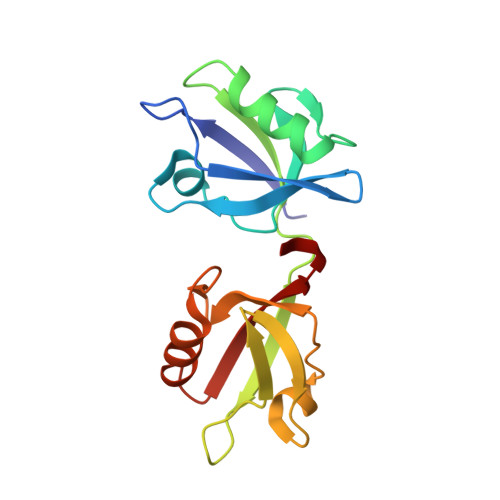
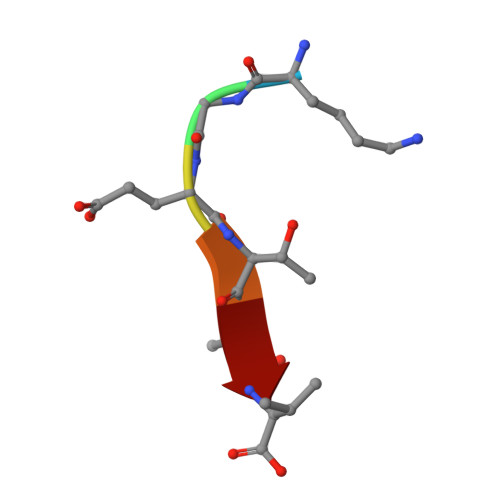
Crystal structure of the syntenin PDZ1 and PDZ2 tandem in complex with the Frizzled 7 C-terminal fragment and PIP2
Egea-Jimenez, A.L., Gallardo, R., Garcia-Pino, A., Ivarsson, Y., Wawrzyniak, A.M., Kashyap, R., Loris, R., Schymkowitz, J., Rousseau, F., Zimmermann, P.To be published.