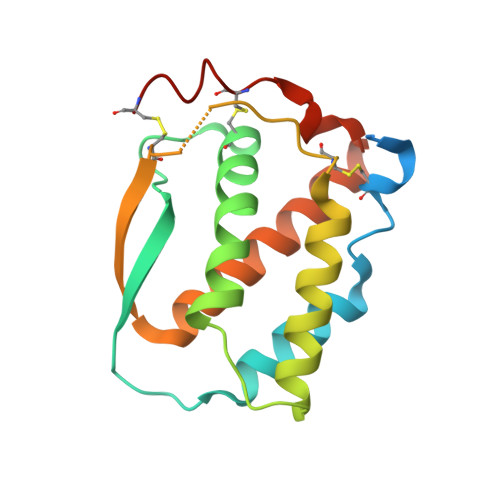
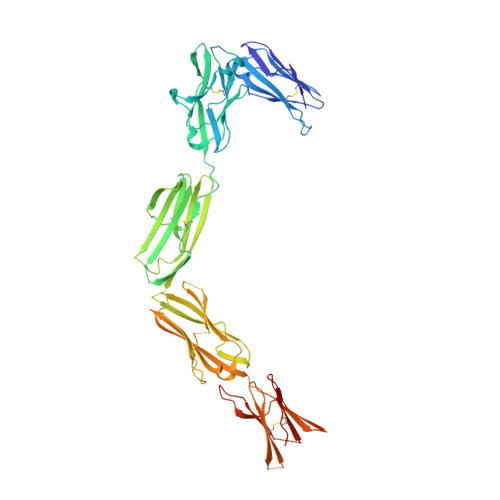Structure and Assembly Mechanism of the Signaling Complex Mediated by Human CSF-1.
Felix, J., De Munck, S., Verstraete, K., Meuris, L., Callewaert, N., Elegheert, J., Savvides, S.N.(2015) Structure 23: 1621-1631
- PubMed: 26235028
- DOI: https://doi.org/10.1016/j.str.2015.06.019
- Primary Citation of Related Structures:
4WRL, 4WRM - PubMed Abstract:
Human colony-stimulating factor 1 receptor (hCSF-1R) is unique among the hematopoietic receptors because it is activated by two distinct cytokines, CSF-1 and interleukin-34 (IL-34). Despite ever-growing insights into the central role of hCSF-1R signaling in innate and adaptive immunity, inflammatory diseases, and cancer, the structural basis of the functional dichotomy of hCSF-1R has remained elusive. Here, we report crystal structures of ternary complexes between hCSF-1 and hCSF-1R, including their complete extracellular assembly, and propose a mechanism for the cooperative human CSF-1:CSF-1R complex that relies on the adoption by dimeric hCSF-1 of an active conformational state and homotypic receptor interactions. Furthermore, we trace the cytokine-binding duality of hCSF-1R to a limited set of conserved interactions mediated by functionally equivalent residues on CSF-1 and IL-34 that play into the geometric requirements of hCSF-1R activation, and map the possible mechanistic consequences of somatic mutations in hCSF-1R associated with cancer.
Organizational Affiliation:
Laboratory for Protein Biochemistry and Biomolecular Engineering (L-ProBE), Department of Biochemistry and Microbiology, Ghent University, 9000 Ghent, Belgium; Unit for Structural Biology, VIB Inflammation Research Center, 9052 Ghent, Belgium.