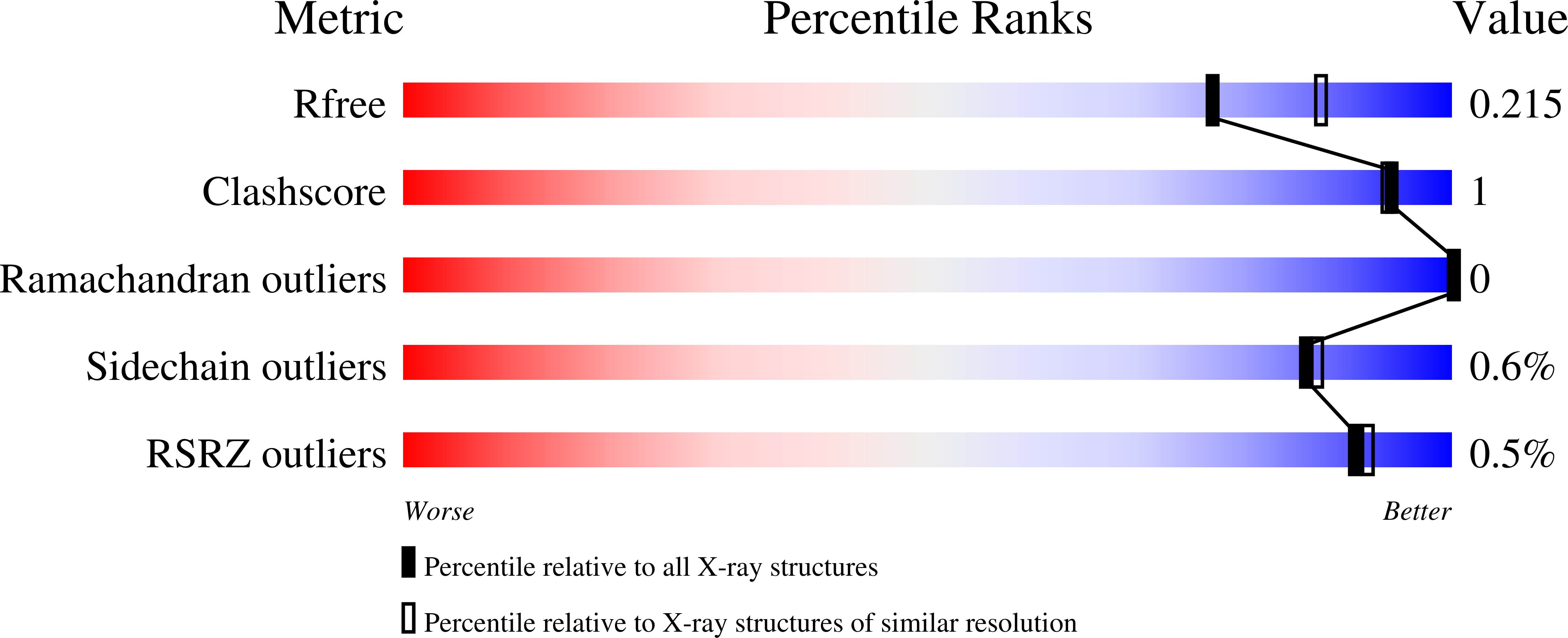
Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Da Fonseca, I., Qureshi, I.A., Mehra-Chaudhary, R., Kizjakina, K., Tanner, J.J., Sobrado, P.(2014) Biochemistry 53: 7794-7804
- PubMed: 25412209
- DOI: https://doi.org/10.1021/bi501008z
- Primary Citation of Related Structures:
4U8I, 4U8J, 4U8K, 4U8L, 4U8M, 4U8N, 4U8O, 4U8P, 4WX1 - PubMed Abstract:
UDP-galactopyranose mutase (UGM) catalyzes the interconversion between UDP-galactopyranose and UDP-galactofuranose. Absent in humans, galactofuranose is found in bacterial and fungal cell walls and is a cell surface virulence factor in protozoan parasites. For these reasons, UGMs are targets for drug discovery. Here, we report a mutagenesis and structural study of the UGMs from Aspergillus fumigatus and Trypanosoma cruzi focused on active site residues that are conserved in eukaryotic UGMs but are absent or different in bacterial UGMs. Kinetic analysis of the variants F66A, Y104A, Q107A, N207A, and Y317A (A. fumigatus numbering) show decreases in k(cat)/K(M) values of 200-1000-fold for the mutase reaction. In contrast, none of the mutations significantly affect the kinetics of enzyme activation by NADPH. These results indicate that the targeted residues are important for promoting the transition state conformation for UDP-galactofuranose formation. Crystal structures of the A. fumigatus mutant enzymes were determined in the presence and absence of UDP to understand the structural consequences of the mutations. The structures suggest important roles for Asn207 in stabilizing the closed active site, and Tyr317 in positioning of the uridine ring. Phe66 and the corresponding residue in Mycobacterium tuberculosis UGM (His68) play a role as the backstop, stabilizing the galactopyranose group for nucleophilic attack. Together, these results provide insight into the essentiality of the targeted residues for realizing maximal catalytic activity and a proposal for how conformational changes that close the active site are temporally related and coupled together.
Organizational Affiliation:
Department of Biochemistry, Virginia Tech , Blacksburg, Virginia 24061, United States.