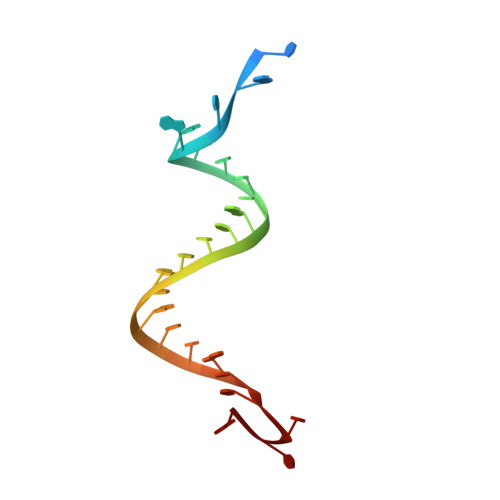
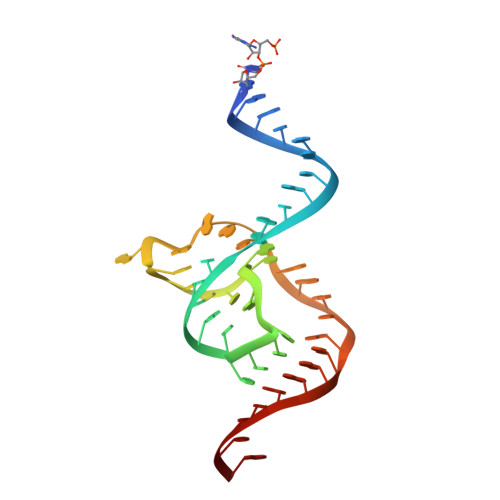
Structural basis for the fast self-cleavage reaction catalyzed by the twister ribozyme.
Eiler, D., Wang, J., Steitz, T.A.(2014) Proc Natl Acad Sci U S A 111: 13028-13033
- PubMed: 25157168
- DOI: https://doi.org/10.1073/pnas.1414571111
- Primary Citation of Related Structures:
4QJD, 4QJH - PubMed Abstract:
Twister is a recently discovered RNA motif that is estimated to have one of the fastest known catalytic rates of any naturally occurring small self-cleaving ribozyme. We determined the 4.1-Å resolution crystal structure of a twister sequence from an organism that has not been cultured in isolation, and it shows an ordered scissile phosphate and nucleotide 5' to the cleavage site. A second crystal structure of twister from Orzyza sativa determined at 3.1-Å resolution exhibits a disordered scissile phosphate and nucleotide 5' to the cleavage site. The core of twister is stabilized by base pairing, a large network of stacking interactions, and two pseudoknots. We observe three nucleotides that appear to mediate catalysis: a guanosine that we propose deprotonates the 2'-hydroxyl of the nucleotide 5' to the cleavage site and a conserved adenosine. We suggest the adenosine neutralizes the negative charge on a nonbridging phosphate oxygen atom at the cleavage site. The active site also positions the labile linkage for in-line nucleophilic attack, and thus twister appears to simultaneously use three strategies proposed for small self-cleaving ribozymes. The twister crystal structures (i) show its global structure, (ii) demonstrate the significance of the double pseudoknot fold, (iii) provide a possible hypothesis for enhanced catalysis, and (iv) illuminate the roles of all 10 highly conserved nucleotides of twister that participate in the formation of its small and stable catalytic pocket.
Organizational Affiliation:
Department of Molecular Biochemistry and Biophysics and.