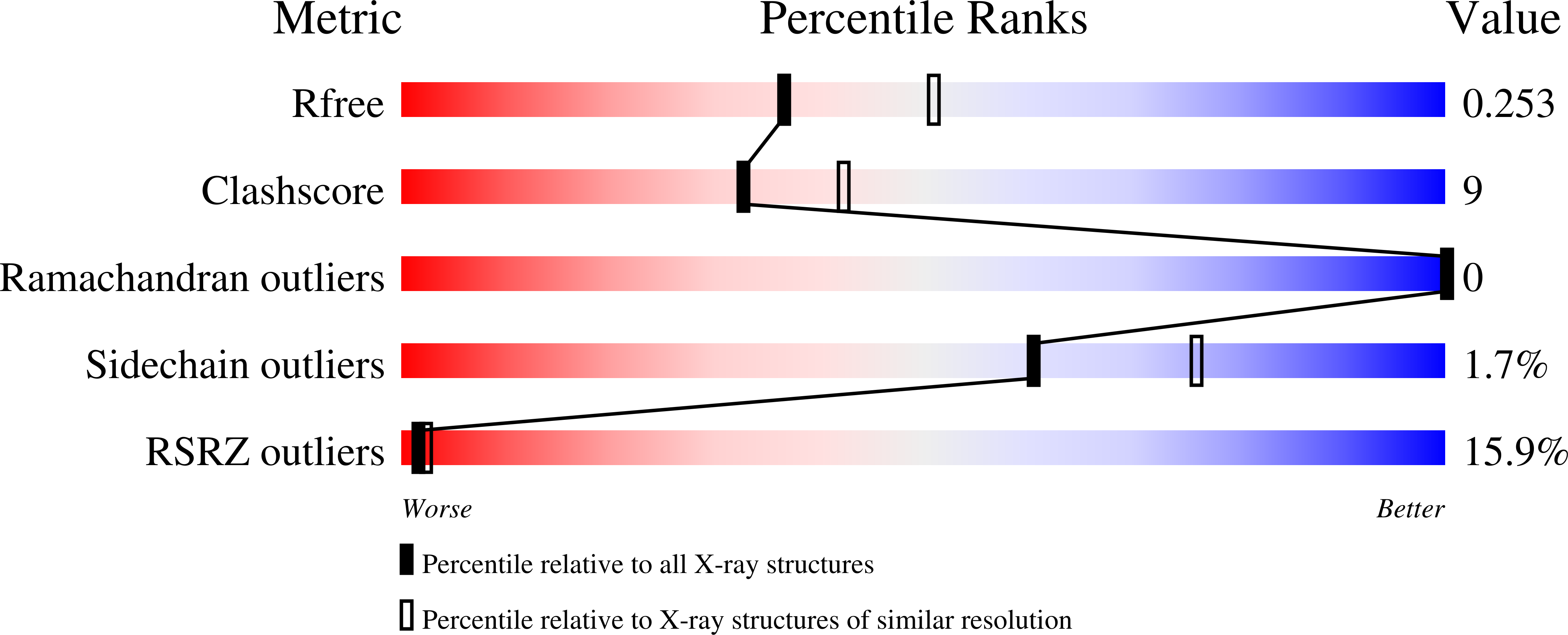
Multiple polymer architectures of human polyhomeotic homolog 3 sterile alpha motif.
Nanyes, D.R., Junco, S.E., Taylor, A.B., Robinson, A.K., Patterson, N.L., Shivarajpur, A., Halloran, J., Hale, S.M., Kaur, Y., Hart, P.J., Kim, C.A.(2014) Proteins 82: 2823-2830
- PubMed: 25044168
- DOI: https://doi.org/10.1002/prot.24645
- Primary Citation of Related Structures:
4PZN, 4PZO - PubMed Abstract:
The self-association of sterile alpha motifs (SAMs) into a helical polymer architecture is a critical functional component of many different and diverse array of proteins. For the Drosophila Polycomb group (PcG) protein Polyhomeotic (Ph), its SAM polymerization serves as the structural foundation to cluster multiple PcG complexes, helping to maintain a silenced chromatin state. Ph SAM shares 64% sequence identity with its human ortholog, PHC3 SAM, and both SAMs polymerize. However, in the context of their larger protein regions, PHC3 SAM forms longer polymers compared with Ph SAM. Motivated to establish the precise structural basis for the differences, if any, between Ph and PHC3 SAM, we determined the crystal structure of the PHC3 SAM polymer. PHC3 SAM uses the same SAM-SAM interaction as the Ph SAM sixfold repeat polymer. Yet, PHC3 SAM polymerizes using just five SAMs per turn of the helical polymer rather than the typical six per turn observed for all SAM polymers reported to date. Structural analysis suggested that malleability of the PHC3 SAM would allow formation of not just the fivefold repeat structure but also possibly others. Indeed, a second PHC3 SAM polymer in a different crystal form forms a sixfold repeat polymer. These results suggest that the polymers formed by PHC3 SAM, and likely others, are dynamic. The functional consequence of the variable PHC3 SAM polymers may be to create different chromatin architectures.
Organizational Affiliation:
Department of Biochemistry, The University of Texas Health Science Center San Antonio, MSC 7760, 7703 Floyd Curl Dr., San Antonio, Texas, 78229.