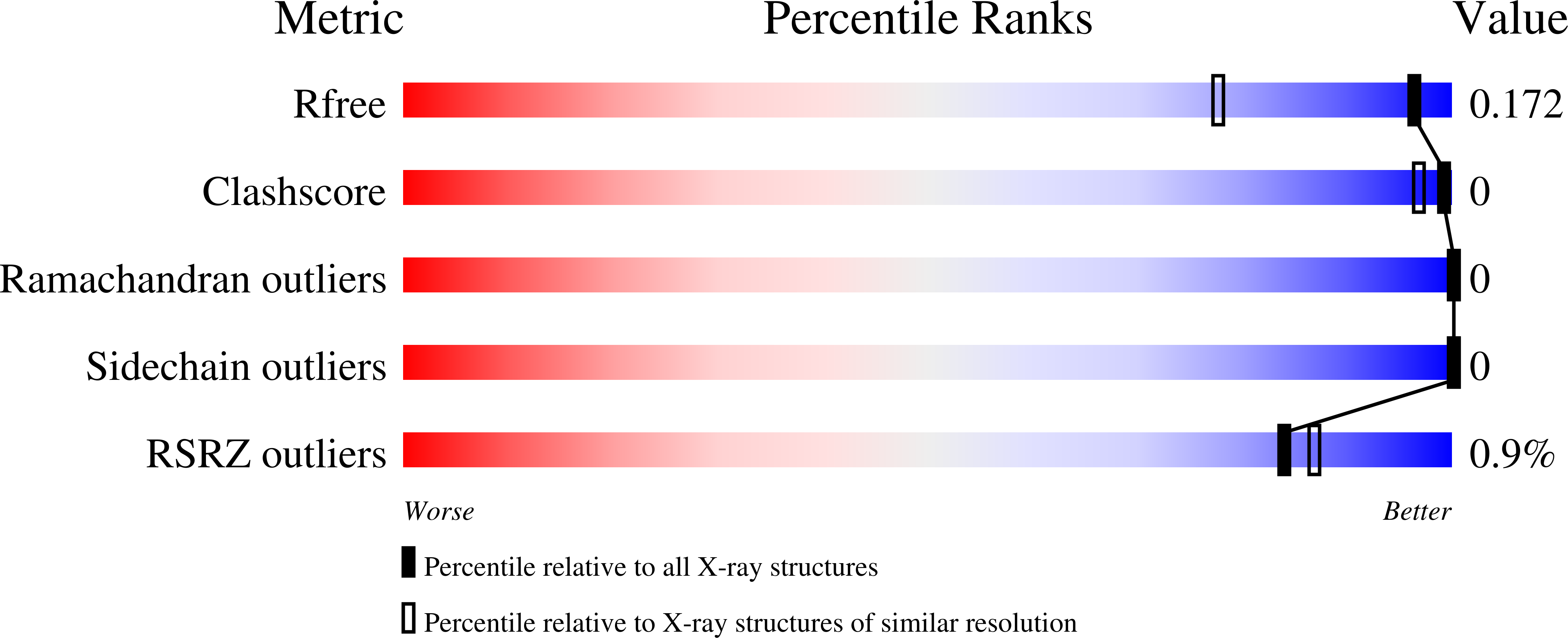
Structural studies suggest a peptidoglycan hydrolase function for the Mycobacterium tuberculosis Tat-secreted protein Rv2525c.
Bellinzoni, M., Haouz, A., Miras, I., Magnet, S., Andre-Leroux, G., Mukherjee, R., Shepard, W., Cole, S.T., Alzari, P.M.(2014) J Struct Biol 188: 156-164
- PubMed: 25260828
- DOI: https://doi.org/10.1016/j.jsb.2014.09.003
- Primary Citation of Related Structures:
4PMN, 4PMO, 4PMQ, 4PMR - PubMed Abstract:
Among the few proteins shown to be secreted by the Tat system in Mycobacterium tuberculosis, Rv2525c is of particular interest, since its gene is conserved in the minimal genome of Mycobacterium leprae. Previous evidence linked this protein to cell wall metabolism and sensitivity to β-lactams. We describe here the crystal structure of Rv2525c that shows a TIM barrel-like fold characteristic of glycoside hydrolases of the GH25 family, which includes prokaryotic and phage-encoded peptidoglycan hydrolases. Structural comparison with other members of this family combined with substrate docking suggest that, although the 'neighbouring group' catalytic mechanism proposed for this family still appears as the most plausible, the identity of residues involved in catalysis in GH25 hydrolases might need to be revised.
Organizational Affiliation:
Institut Pasteur, Unité de Microbiologie Structurale and CNRS-UMR3528, 25 rue du Dr. Roux, 75724 Paris Cedex 15, France. Electronic address: marco.bellinzoni@pasteur.fr.