Synthetic Inositol Phosphate Analogs Reveal that PPIP5K2 Has a Surface-Mounted Substrate Capture Site that Is a Target for Drug Discovery.
Wang, H., Godage, H.Y., Riley, A.M., Weaver, J.D., Shears, S.B., Potter, B.V.(2014) Chem Biol 21: 689-699
- PubMed: 24768307
- DOI: https://doi.org/10.1016/j.chembiol.2014.03.009
- Primary Citation of Related Structures:
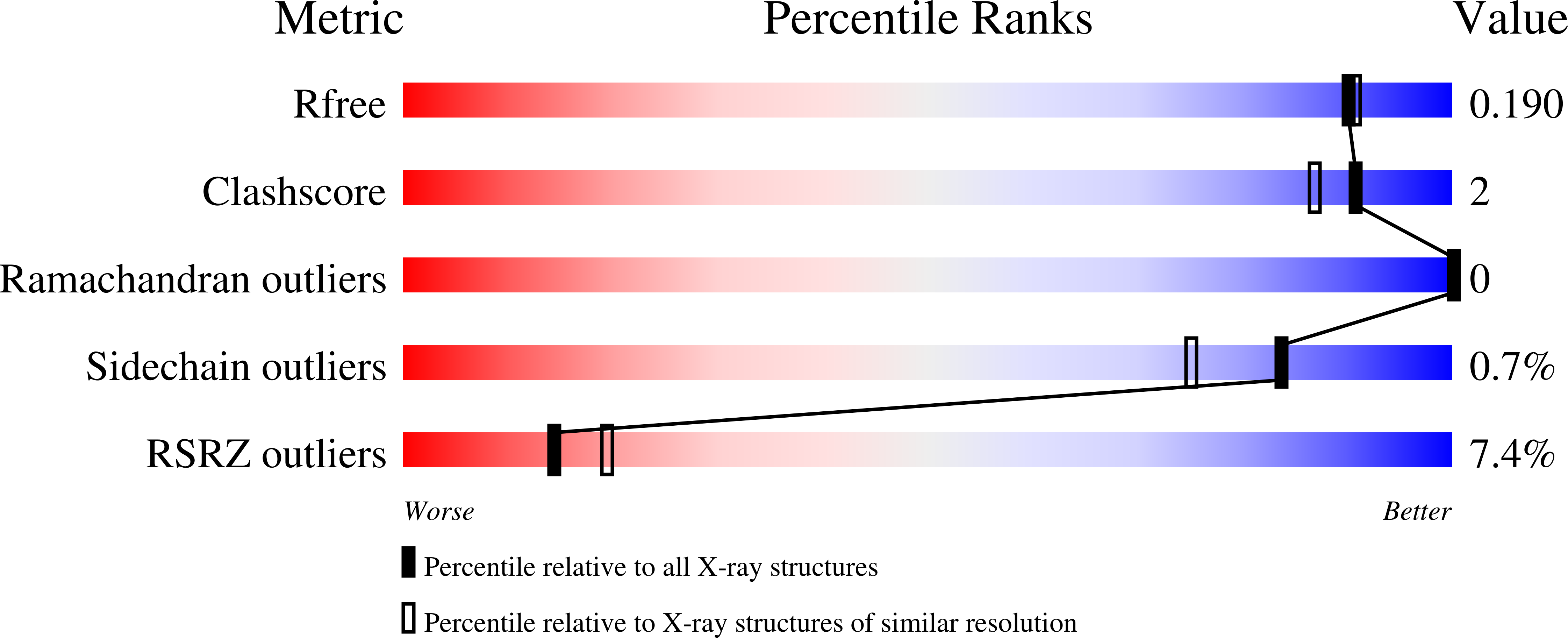
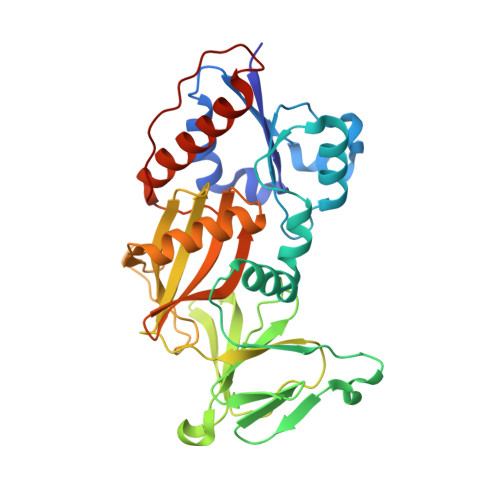
4NZM, 4NZN, 4NZO - PubMed Abstract:
Diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) is one of the mammalian PPIP5K isoforms responsible for synthesis of diphosphoinositol polyphosphates (inositol pyrophosphates; PP-InsPs), regulatory molecules that function at the interface of cell signaling and organismic homeostasis. The development of drugs that inhibit PPIP5K2 could have both experimental and therapeutic applications. Here, we describe a synthetic strategy for producing naturally occurring 5-PP-InsP4, as well as several inositol polyphosphate analogs, and we study their interactions with PPIP5K2 using biochemical and structural approaches. These experiments uncover an additional ligand-binding site on the surface of PPIP5K2, adjacent to the catalytic pocket. This site facilitates substrate capture from the bulk phase, prior to transfer into the catalytic pocket. In addition to demonstrating a "catch-and-pass" reaction mechanism in a small molecule kinase, we demonstrate that binding of our analogs to the substrate capture site inhibits PPIP5K2. This work suggests that the substrate-binding site offers new opportunities for targeted drug design.
Organizational Affiliation:
Inositol Signaling Group, Laboratory of Signal Transduction, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.