Streptococcus sanguinis Class Ib Ribonucleotide Reductase: HIGH ACTIVITY WITH BOTH IRON AND MANGANESE COFACTORS AND STRUCTURAL INSIGHTS.
Makhlynets, O., Boal, A.K., Rhodes, D.V., Kitten, T., Rosenzweig, A.C., Stubbe, J.(2014) J Biol Chem 289: 6259-6272
- PubMed: 24381172
- DOI: https://doi.org/10.1074/jbc.M113.533554
- Primary Citation of Related Structures:
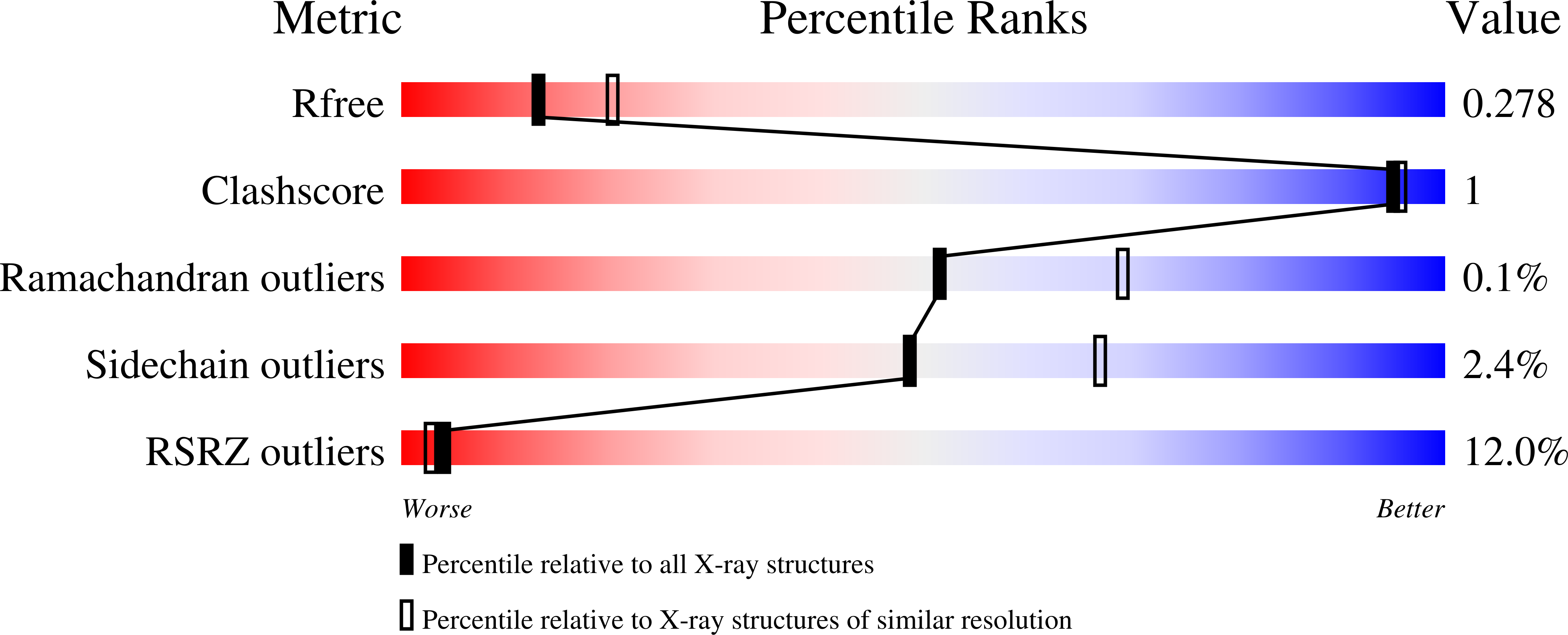
4N82, 4N83 - PubMed Abstract:
Streptococcus sanguinis is a causative agent of infective endocarditis. Deletion of SsaB, a manganese transporter, drastically reduces S. sanguinis virulence. Many pathogenic organisms require class Ib ribonucleotide reductase (RNR) to catalyze the conversion of nucleotides to deoxynucleotides under aerobic conditions, and recent studies demonstrate that this enzyme uses a dimanganese-tyrosyl radical (Mn(III)2-Y(•)) cofactor in vivo. The proteins required for S. sanguinis ribonucleotide reduction (NrdE and NrdF, α and β subunits of RNR; NrdH and TrxR, a glutaredoxin-like thioredoxin and a thioredoxin reductase; and NrdI, a flavodoxin essential for assembly of the RNR metallo-cofactor) have been identified and characterized. Apo-NrdF with Fe(II) and O2 can self-assemble a diferric-tyrosyl radical (Fe(III)2-Y(•)) cofactor (1.2 Y(•)/β2) and with the help of NrdI can assemble a Mn(III)2-Y(•) cofactor (0.9 Y(•)/β2). The activity of RNR with its endogenous reductants, NrdH and TrxR, is 5,000 and 1,500 units/mg for the Mn- and Fe-NrdFs (Fe-loaded NrdF), respectively. X-ray structures of S. sanguinis NrdIox and Mn(II)2-NrdF are reported and provide a possible rationale for the weak affinity (2.9 μM) between them. These streptococcal proteins form a structurally distinct subclass relative to other Ib proteins with unique features likely important in cluster assembly, including a long and negatively charged loop near the NrdI flavin and a bulky residue (Thr) at a constriction in the oxidant channel to the NrdI interface. These studies set the stage for identifying the active form of S. sanguinis class Ib RNR in an animal model for infective endocarditis and establishing whether the manganese requirement for pathogenesis is associated with RNR.
Organizational Affiliation:
From the Departments of Chemistry and.