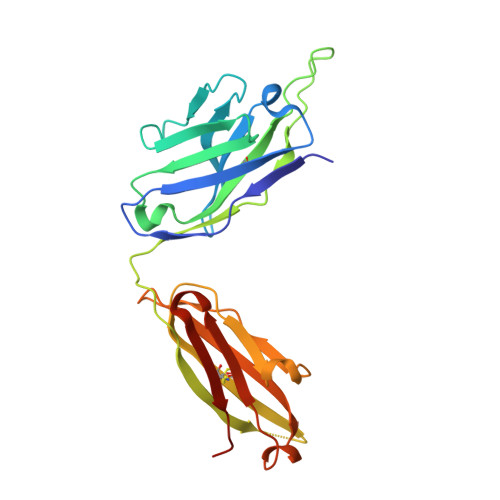
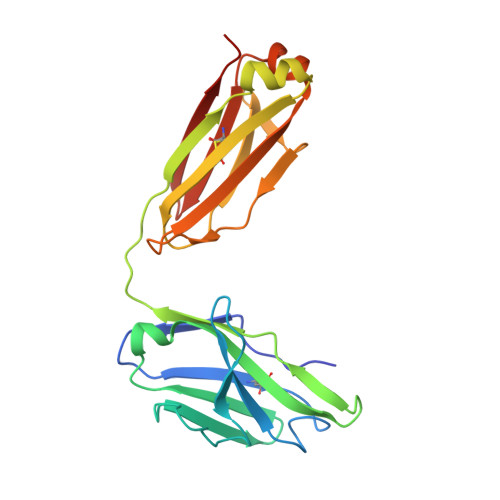
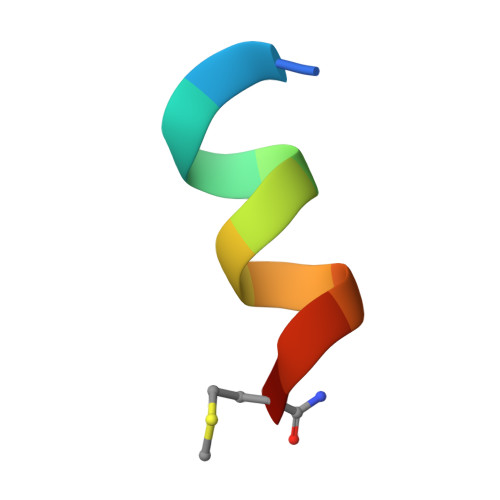Structure of Hepatitis C Virus Envelope Glycoprotein E1 Antigenic Site 314-324 in Complex with Antibody IGH526.
Kong, L., Kadam, R.U., Giang, E., Ruwona, T.B., Nieusma, T., Culhane, J.C., Stanfield, R.L., Dawson, P.E., Wilson, I.A., Law, M.(2015) J Mol Biol 427: 2617-2628
- PubMed: 26135247
- DOI: https://doi.org/10.1016/j.jmb.2015.06.012
- Primary Citation of Related Structures:
4N0Y - PubMed Abstract:
Hepatitis C virus (HCV) is a positive-strand RNA virus within the Flaviviridae family. The viral "spike" of HCV is formed by two envelope glycoproteins, E1 and E2, which together mediate viral entry by engaging host receptors and undergoing conformational changes to facilitate membrane fusion. While E2 can be readily produced in the absence of E1, E1 cannot be expressed without E2 and few reagents, including monoclonal antibodies (mAbs), are available for study of this essential HCV glycoprotein. A human mAb to E1, IGH526, was previously reported to cross-neutralize different HCV isolates, and therefore, we sought to further characterize the IGH526 neutralizing epitope to obtain information for vaccine design. We found that mAb IGH526 bound to a discontinuous epitope, but with a major component corresponding to E1 residues 314-324. The crystal structure of IGH526 Fab with this E1 glycopeptide at 1.75Å resolution revealed that the antibody binds to one face of an α-helical peptide. Single mutations on the helix substantially lowered IGH526 binding but did not affect neutralization, indicating either that multiple mutations are required or that additional regions are recognized by the antibody in the context of the membrane-associated envelope oligomer. Molecular dynamics simulations indicate that the free peptide is flexible in solution, suggesting that it requires stabilization for use as a candidate vaccine immunogen.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.