Molecular basis of ligand recognition by OASS from E. histolytica: insights from structural and molecular dynamics simulation studies
Raj, I., Mazumder, M., Gourinath, S.(2013) Biochim Biophys Acta 1830: 4573-4583
- PubMed: 23747298
- DOI: https://doi.org/10.1016/j.bbagen.2013.05.041
- Primary Citation of Related Structures:
4IL5, 4JBL, 4JBN - PubMed Abstract:
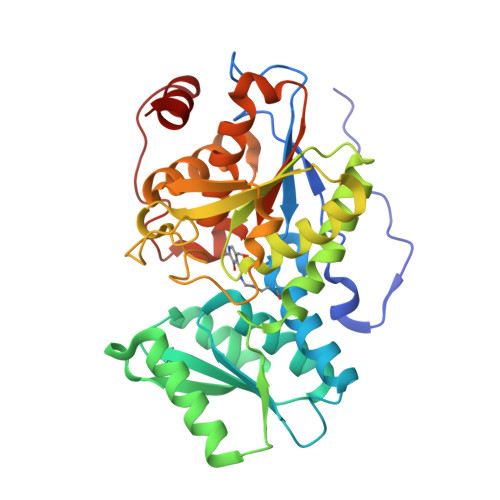
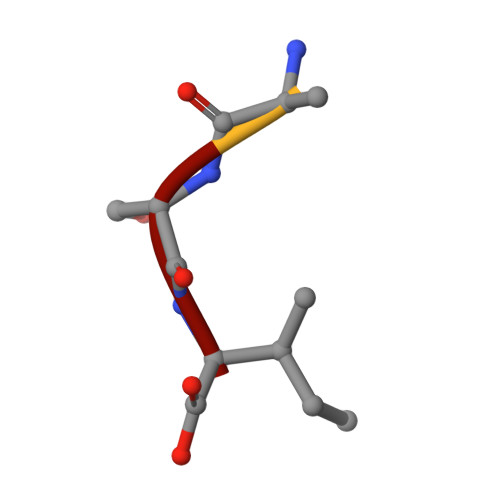
O-acetyl serine sulfhydrylase (OASS) is a pyridoxal phosphate (PLP) dependent enzyme catalyzing the last step of the cysteine biosynthetic pathway. Here we analyze and investigate the factors responsible for recognition and different conformational changes accompanying the binding of various ligands to OASS. X ray crystallography was used to determine the structures of OASS from Entamoeba histolytica in complex with methionine (substrate analog), isoleucine (inhibitor) and an inhibitory tetra-peptide to 2.00Å, 2.03Å and 1.87Å resolutions, respectively. Molecular dynamics simulations were used to investigate the reasons responsible for the extent of domain movement and cleft closure of the enzyme in presence of different ligands. Here we report for the first time an OASS-methionine structure with an unmutated catalytic lysine at the active site. This is also the first OASS structure with a closed active site lacking external aldimine formation. The OASS-isoleucine structure shows the active site cleft in open state. Molecular dynamics studies indicate that cofactor PLP, N88 and G192 form a triad of energy contributors to close the active site upon ligand binding and orientation of the Schiff base forming nitrogen of the ligand is critical for this interaction. Methionine proves to be a better binder to OASS than isoleucine. The β branching of isoleucine does not allow it to reorient itself in suitable conformation near PLP to cause active site closure. Our findings have important implications in designing better inhibitors against OASS across all pathogenic microbial species.
Organizational Affiliation:
School of Life Sciences, Jawaharlal Nehru University, New Delhi, India.