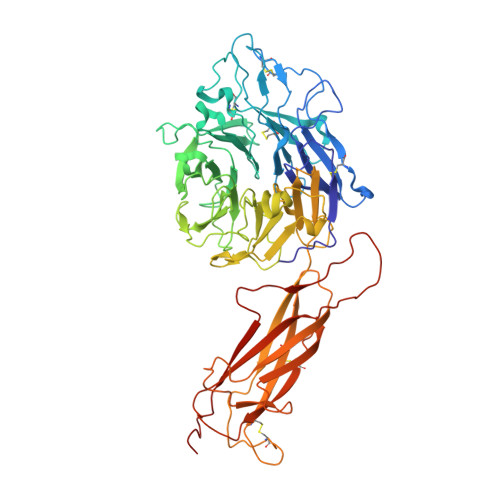
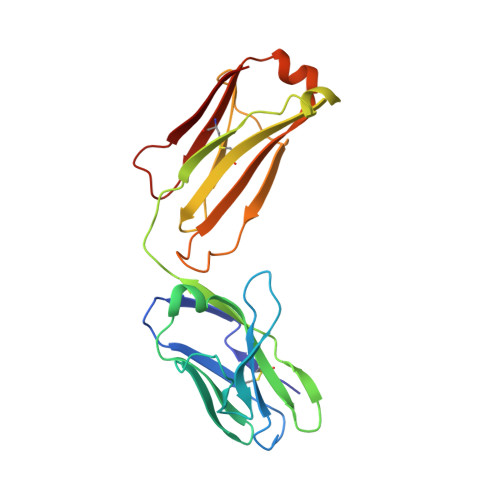
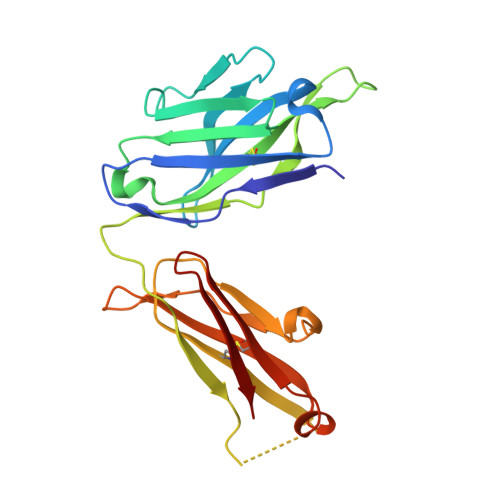
How natalizumab binds and antagonizes alpha 4 integrins.
Yu, Y., Schurpf, T., Springer, T.A.(2013) J Biol Chem 288: 32314-32325
- PubMed: 24047894
- DOI: https://doi.org/10.1074/jbc.M113.501668
- Primary Citation of Related Structures:
4IRZ - PubMed Abstract:
Natalizumab antibody to α4-integrins is used in therapy of multiple sclerosis and Crohn's disease. A crystal structure of the Fab bound to an α4 integrin β-propeller and thigh domain fragment shows that natalizumab recognizes human-mouse differences on the circumference of the β-propeller domain. The epitope is adjacent to but outside of a ligand-binding groove formed at the interface with the β-subunit βI domain and shows no difference in structure when bound to Fab. Competition between Fab and the ligand vascular cell adhesion molecule (VCAM) for binding to cell surface α4β1 shows noncompetitive antagonism. In agreement, VCAM docking models suggest that binding of domain 1 of VCAM to α4-integrins is unimpeded by the Fab, and that bound Fab requires a change in orientation between domains 1 and 2 of VCAM for binding to α4β1. Mapping of species-specific differences onto α4β1 and α4β7 shows that their ligand-binding sites are highly conserved. Skewing away from these conserved regions of the epitopes recognized by current therapeutic function-blocking antibodies has resulted in previously unanticipated mechanisms of action.
Organizational Affiliation:
From the Program in Cellular and Molecular Medicine, Children's Hospital Boston and Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, Massachusetts 02115.