A dual anchoring strategy for the localization and activation of artificial metalloenzymes based on the biotin-streptavidin technology.
Zimbron, J.M., Heinisch, T., Schmid, M., Hamels, D., Nogueira, E.S., Schirmer, T., Ward, T.R.(2013) J Am Chem Soc 135: 5384-5388
- PubMed: 23496309
- DOI: https://doi.org/10.1021/ja309974s
- Primary Citation of Related Structures:
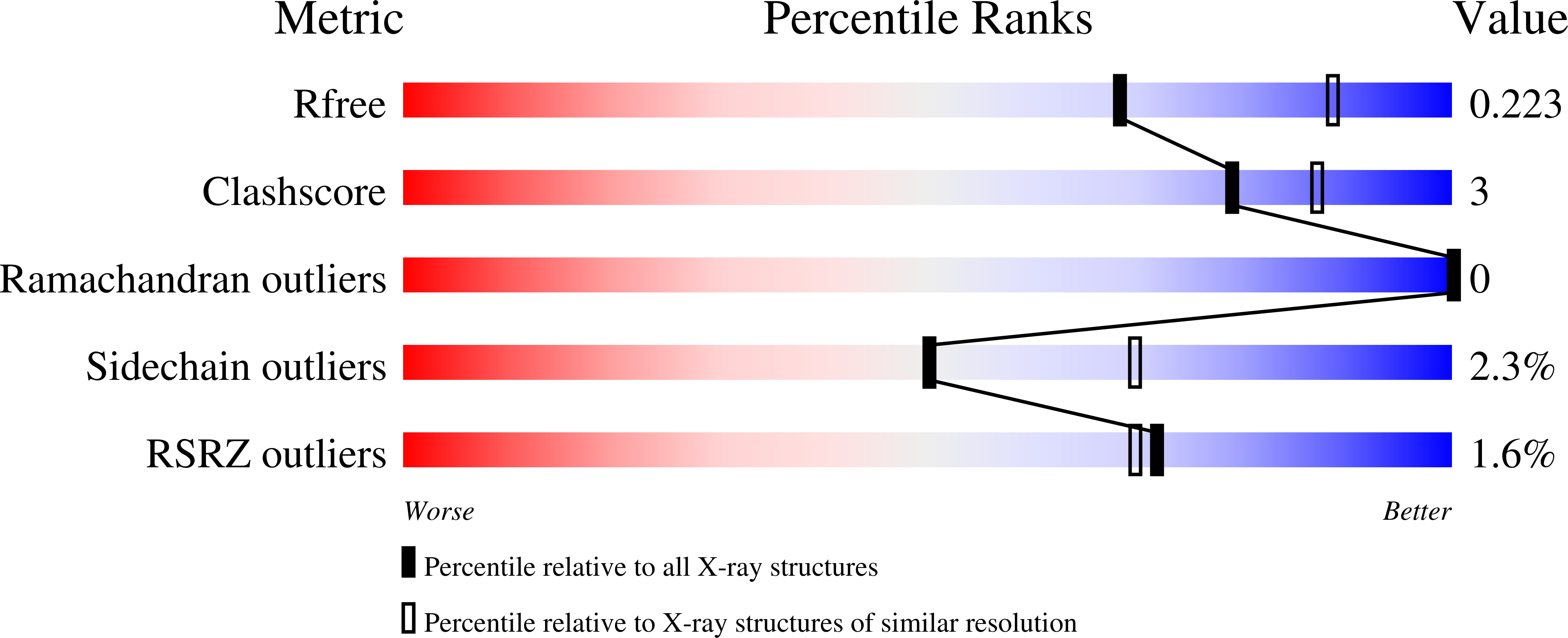
4GJS, 4GJV - PubMed Abstract:
Artificial metalloenzymes result from anchoring an active catalyst within a protein environment. Toward this goal, various localization strategies have been pursued: covalent, supramolecular, or dative anchoring. Herein we show that introduction of a suitably positioned histidine residue contributes to firmly anchor, via a dative bond, a biotinylated rhodium piano stool complex within streptavidin. The in silico design of the artificial metalloenzyme was confirmed by X-ray crystallography. The resulting artificial metalloenzyme displays significantly improved catalytic performance, both in terms of activity and selectivity in the transfer hydrogenation of imines. Depending on the position of the histidine residue, both enantiomers of the salsolidine product can be obtained.
Organizational Affiliation:
Biozentrum and Department of Chemistry, University of Basel, CH-4056 Basel, Switzerland.