Matured structure of anti-HIV lectin actinohivin in complex with 1,2-mannobiose
Hoque, M.M., Suzuki, K., Tsunoda, M., Jiang, J., Zhang, F., Takahashi, A., Naomi, O., Zhang, X., Sekiguchi, T., Tanaka, H., Omura, S., Takenaka, A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Actinohivin | 114 | actinomycete K97-0003 | Mutation(s): 0 | 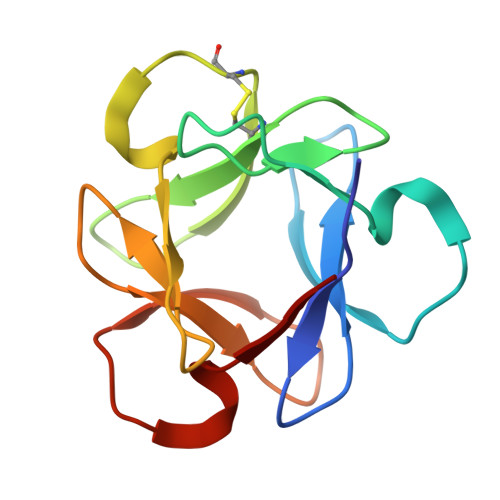 | |
UniProt | |||||
Find proteins for Q9KWN0 (Actinomycete sp. (strain K97-0003)) Explore Q9KWN0 Go to UniProtKB: Q9KWN0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9KWN0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CCN Query on CCN | E [auth A], F [auth A] | ACETONITRILE C2 H3 N WEVYAHXRMPXWCK-UHFFFAOYSA-N |  | ||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900111 Query on PRD_900111 | B, C, D | 2alpha-alpha-mannobiose | Oligosaccharide / Metabolism |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 27.84 | α = 90 |
| b = 66.84 | β = 90 |
| c = 67.06 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| AMoRE | phasing |
| REFMAC | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |