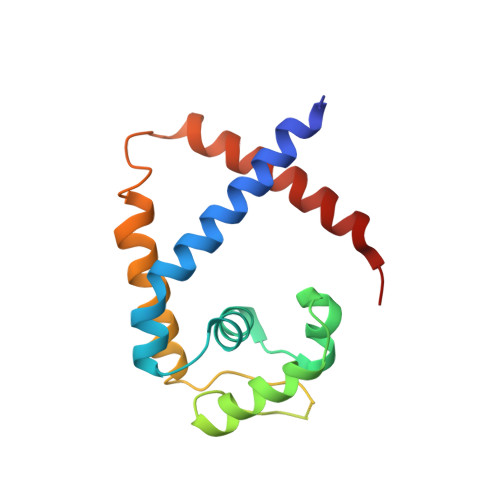
Functional studies of ssDNA binding ability of MarR family protein TcaR from Staphylococcus epidermidis.
Chang, Y.M., Chen, C.K.-M., Chang, Y.C., Jeng, W.Y., Hou, M.H., Wang, A.H.-J.(2012) PLoS One 7: e45665-e45665
- PubMed: 23029170
- DOI: https://doi.org/10.1371/journal.pone.0045665
- Primary Citation of Related Structures:
4EJT - PubMed Abstract:
The negative transcription regulator of the ica locus, TcaR, regulates proteins involved in the biosynthesis of poly-N-acetylglucosamine (PNAG). Absence of TcaR increases PNAG production and promotes biofilm formation in Staphylococci. Previously, the 3D structure of TcaR in its apo form and its complex structure with several antibiotics have been analyzed. However, the detailed mechanism of multiple antibiotic resistance regulator (MarR) family proteins such as TcaR is unclear and only restricted on the binding ability of double-strand DNA (dsDNA). Here we show by electrophoretic mobility shift assay (EMSA), electron microscopy (EM), circular dichroism (CD), and Biacore analysis that TcaR can interact strongly with single-stranded DNA (ssDNA), thereby identifying a new role in MarR family proteins. Moreover, we show that TcaR preferentially binds 33-mer ssDNA over double-stranded DNA and inhibits viral ssDNA replication. In contrast, such ssDNA binding properties were not observed for other MarR family protein and TetR family protein, suggesting that the results from our studies are not an artifact due to simple charge interactions between TcaR and ssDNA. Overall, these results suggest a novel role for TcaR in regulation of DNA replication. We anticipate that the results of this work will extend our understanding of MarR family protein and broaden the development of new therapeutic strategies for Staphylococci.
Organizational Affiliation:
Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan.