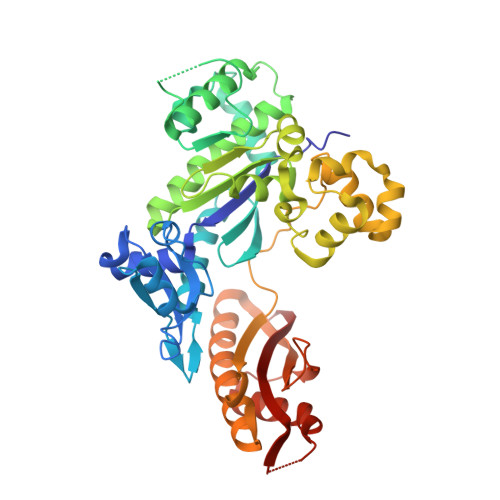
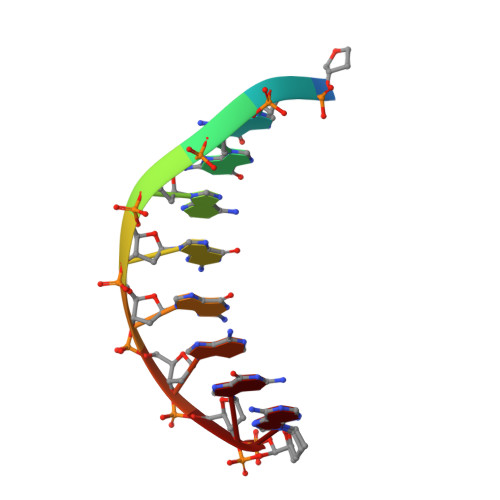
Structural basis for cisplatin DNA damage tolerance by human polymerase {eta} during cancer chemotherapy.
Ummat, A., Rechkoblit, O., Jain, R., Roy Choudhury, J., Johnson, R.E., Silverstein, T.D., Buku, A., Lone, S., Prakash, L., Prakash, S., Aggarwal, A.K.(2012) Nat Struct Mol Biol 19: 628-632
- PubMed: 22562137
- DOI: https://doi.org/10.1038/nsmb.2295
- Primary Citation of Related Structures:
4EEY - PubMed Abstract:
A major clinical problem in the use of cisplatin to treat cancers is tumor resistance. DNA polymerase η (Pol-η) is a crucial polymerase that allows cancer cells to cope with the cisplatin-DNA adducts that are formed during chemotherapy. We present here a structure of human Pol-η inserting deoxycytidine triphosphate (dCTP) opposite a cisplatin intrastrand cross-link (PtGpG). We show that the specificity of human Pol-η for PtGpG derives from an active site that is open to permit Watson-Crick geometry of the nascent PtGpG-dCTP base pair and to accommodate the lesion without steric hindrance. This specificity is augmented by the residues Gln38 and Ser62, which interact with PtGpG, and Arg61, which interacts with the incoming dCTP. Collectively, the structure provides a basis for understanding how Pol-η in human cells can tolerate the DNA damage caused by cisplatin chemotherapy and offers a framework for the design of inhibitors in cancer therapy.
Organizational Affiliation:
Department of Structural and Chemical Biology, Mount Sinai School of Medicine, New York, New York, USA.