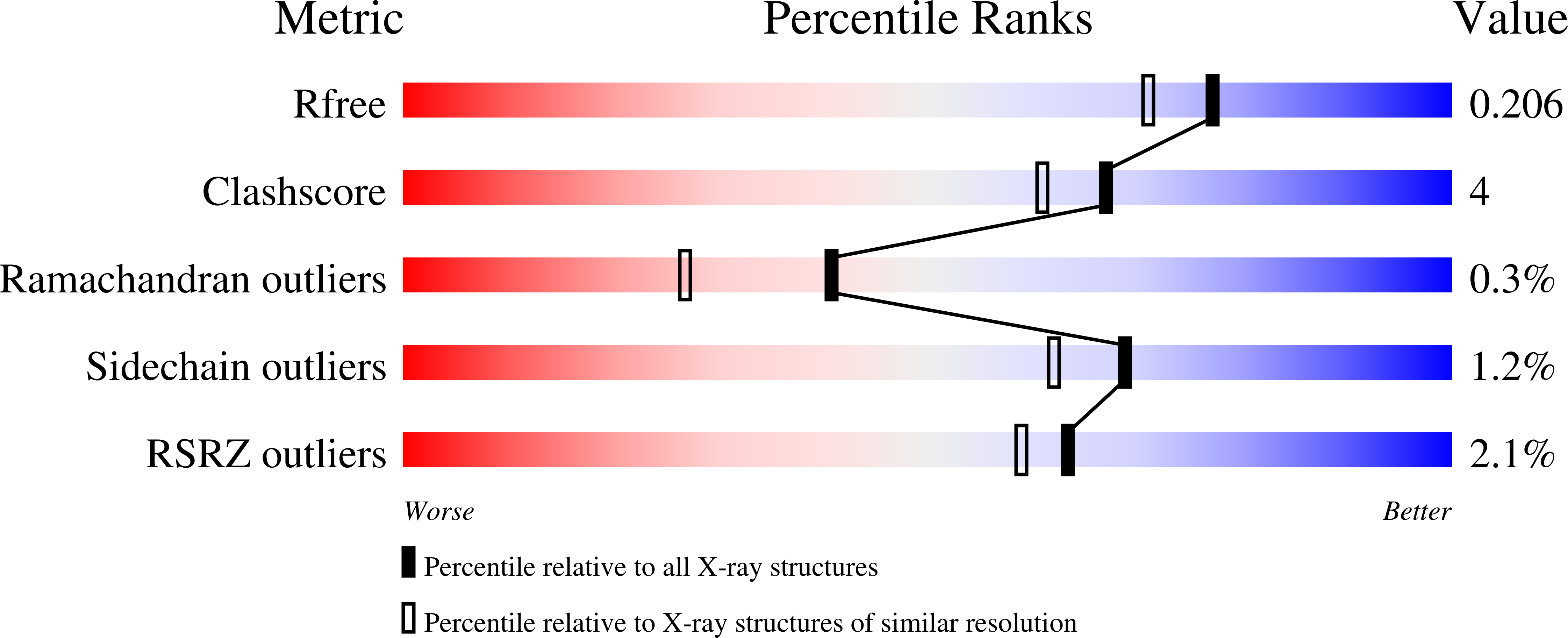
Crystal structure of Putative dehydratase protein from Salmonella enterica subsp. enterica serovar Typhimurium (Salmonella typhimurium)
Malashkevich, V.N., Bhosle, R., Toro, R., Hillerich, B., Gizzi, A., Garforth, S., Kar, A., Chan, M.K., Lafluer, J., Patel, H., Matikainen, B., Chamala, S., Lim, S., Celikgil, A., Villegas, G., Evans, B., Zenchek, W., Love, J., Fiser, A., Khafizov, K., Seidel, R., Bonanno, J.B., Almo, S.C.To be published.