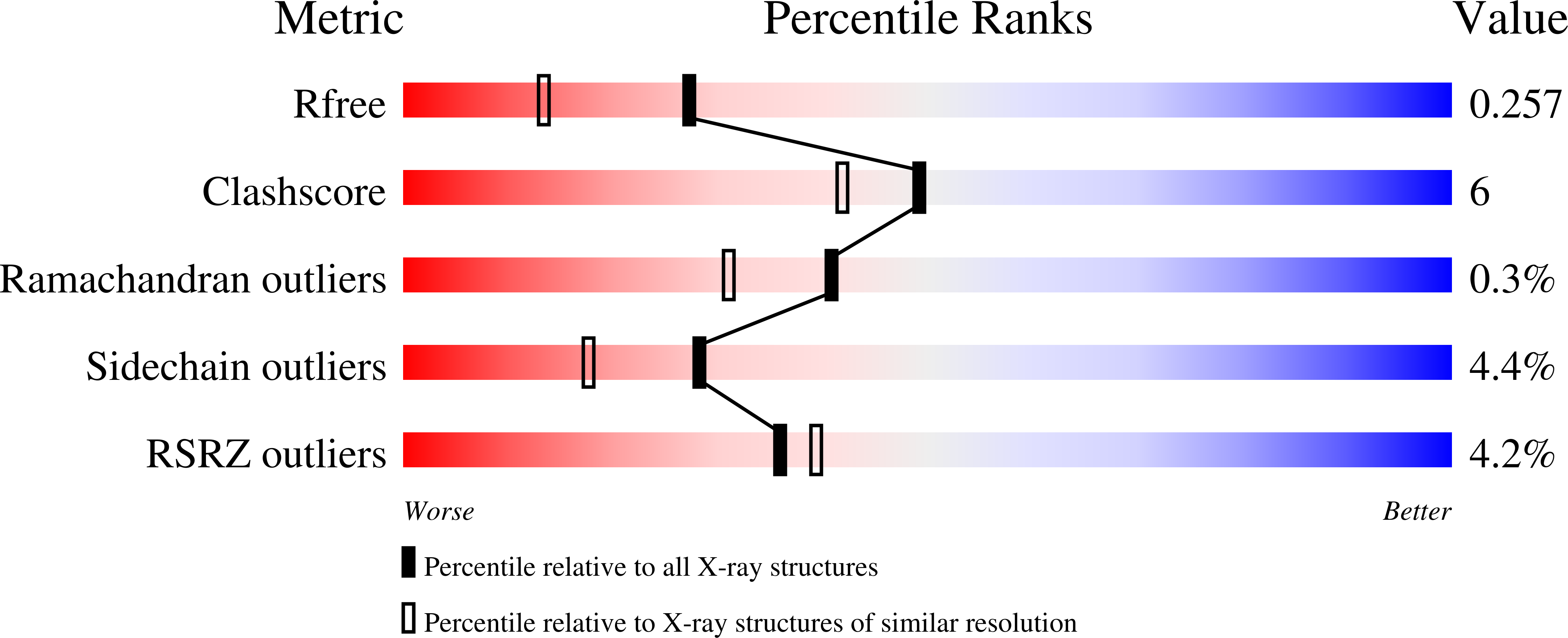
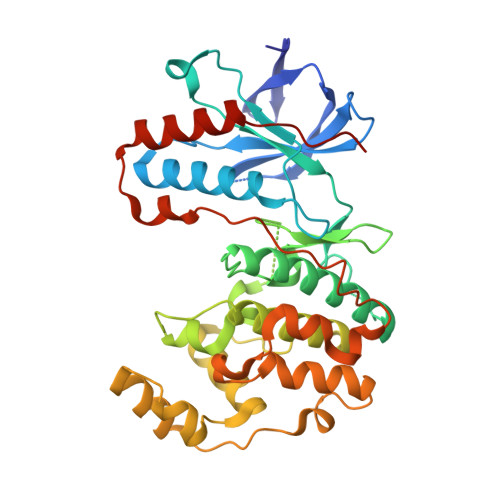
Fluorophore labeled kinase detects ligands that bind within the MAPK insert of p38alpha kinase.
Getlik, M., Simard, J.R., Termathe, M., Grutter, C., Rabiller, M., van Otterlo, W.A., Rauh, D.(2012) PLoS One 7: e39713-e39713
- PubMed: 22768308
- DOI: https://doi.org/10.1371/journal.pone.0039713
- Primary Citation of Related Structures:
4DLI, 4DLJ - PubMed Abstract:
The vast majority of small molecules known to modulate kinase activity, target the highly conserved ATP-pocket. Consequently, such ligands are often less specific and in case of inhibitors, this leads to the inhibition of multiple kinases. Thus, selective modulation of kinase function remains a major hurdle. One of the next great challenges in kinase research is the identification of ligands which bind to less conserved sites and target the non-catalytic functions of protein kinases. However, approaches that allow for the unambiguous identification of molecules that bind to these less conserved sites are few in number. We have previously reported the use of fluorescent labels in kinases (FLiK) to develop direct kinase binding assays that exclusively detect ligands which stabilize inactive (DFG-out) kinase conformations. Here, we present the successful application of the FLiK approach to develop a high-throughput binding assay capable of directly monitoring ligand binding to a remote site within the MAPK insert of p38α mitogen-activated protein kinase (MAPK). Guided by the crystal structure of an initially identified hit molecule in complex with p38α, we developed a tight binding ligand which may serve as an ideal starting point for further investigations of the biological function of the MAPK insert in regulating the p38α signaling pathway.
Organizational Affiliation:
Chemical Genomics Centre of the Max Planck Society, Dortmund, Germany.