Bub3 Reads Phosphorylated Melt Repeats to Promote Spindle Assembly Checkpoint Signaling
Primorac, I., Weir, J.R., Chiroli, E., Gross, F., Hoffmann, I., Van Gerwen, S., Ciliberto, A., Musacchio, A.(2013) Elife 2: 01030
- PubMed: 24066227
- DOI: https://doi.org/10.7554/eLife.01030
- Primary Citation of Related Structures:
4BL0 - PubMed Abstract:
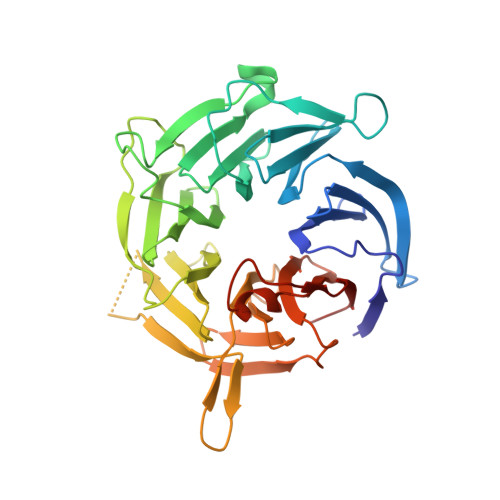
Regulation of macromolecular interactions by phosphorylation is crucial in signaling networks. In the spindle assembly checkpoint (SAC), which enables errorless chromosome segregation, phosphorylation promotes recruitment of SAC proteins to tensionless kinetochores. The SAC kinase Mps1 phosphorylates multiple Met-Glu-Leu-Thr (MELT) motifs on the kinetochore subunit Spc105/Knl1. The phosphorylated MELT motifs (MELT(P)) then promote recruitment of downstream signaling components. How MELT(P) motifs are recognized is unclear. In this study, we report that Bub3, a 7-bladed β-propeller, is the MELT(P) reader. It contains an exceptionally well-conserved interface that docks the MELT(P) sequence on the side of the β-propeller in a previously unknown binding mode. Mutations targeting the Bub3 interface prevent kinetochore recruitment of the SAC kinase Bub1. Crucially, they also cause a checkpoint defect, showing that recognition of phosphorylated targets by Bub3 is required for checkpoint signaling. Our data provide the first detailed mechanistic insight into how phosphorylation promotes recruitment of checkpoint proteins to kinetochores. DOI:http://dx.doi.org/10.7554/eLife.01030.001.
Organizational Affiliation:
Department of Mechanistic Cell Biology , Max Planck Institute of Molecular Physiology , Dortmund , Germany.