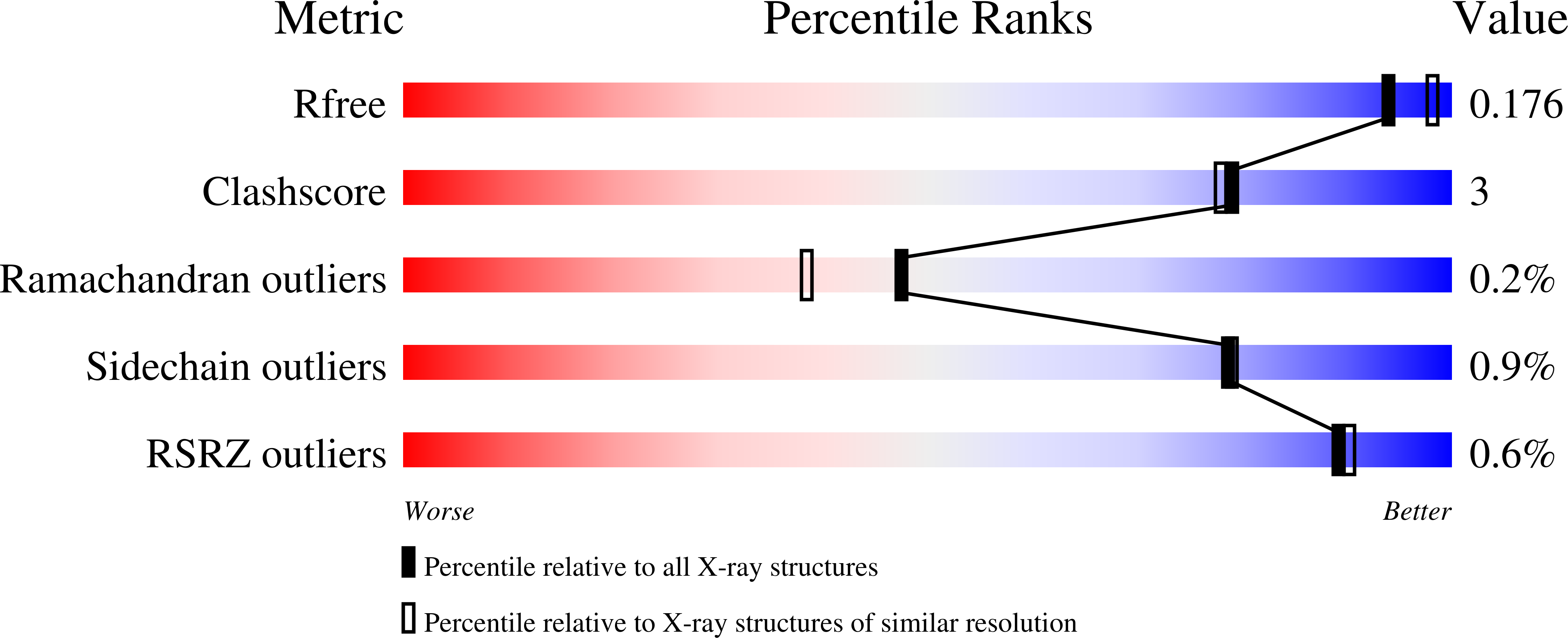
Engineering Methylaspartate Ammonia Lyase for the Asymmetric Synthesis of Unnatural Amino Acids.
Raj, H., Szymanski, W., De Villiers, J., Rozeboom, H.J., Veetil, V.P., Reis, C.R., De Villiers, M., Dekker, F.J., De Wildeman, S., Quax, W.J., Thunnissen, A.M.W.H., Feringa, B.L., Janssen, D.B., Poelarends, G.J.(2012) Nat Chem 4: 478
- PubMed: 22614383
- DOI: https://doi.org/10.1038/nchem.1338
- Primary Citation of Related Structures:
3ZVH, 3ZVI - PubMed Abstract:
The redesign of enzymes to produce catalysts for a predefined transformation remains a major challenge in protein engineering. Here, we describe the structure-based engineering of methylaspartate ammonia lyase (which in nature catalyses the conversion of 3-methylaspartate to ammonia and 2-methylfumarate) to accept a variety of substituted amines and fumarates and catalyse the asymmetric synthesis of aspartic acid derivatives. We obtained two single-active-site mutants, one exhibiting a wide nucleophile scope including structurally diverse linear and cyclic alkylamines and one with broad electrophile scope including fumarate derivatives with alkyl, aryl, alkoxy, aryloxy, alkylthio and arylthio substituents at the C2 position. Both mutants have an enlarged active site that accommodates the new substrates while retaining the high stereo- and regioselectivity of the wild-type enzyme. As an example, we demonstrate a highly enantio- and diastereoselective synthesis of threo-3-benzyloxyaspartate (an important inhibitor of neuronal excitatory glutamate transporters in the brain).
Organizational Affiliation:
Department of Pharmaceutical Biology, Groningen Research Institute of Pharmacy, University of Groningen, Antonius Deusinglaan 1, 9713 AV Groningen, The Netherlands.