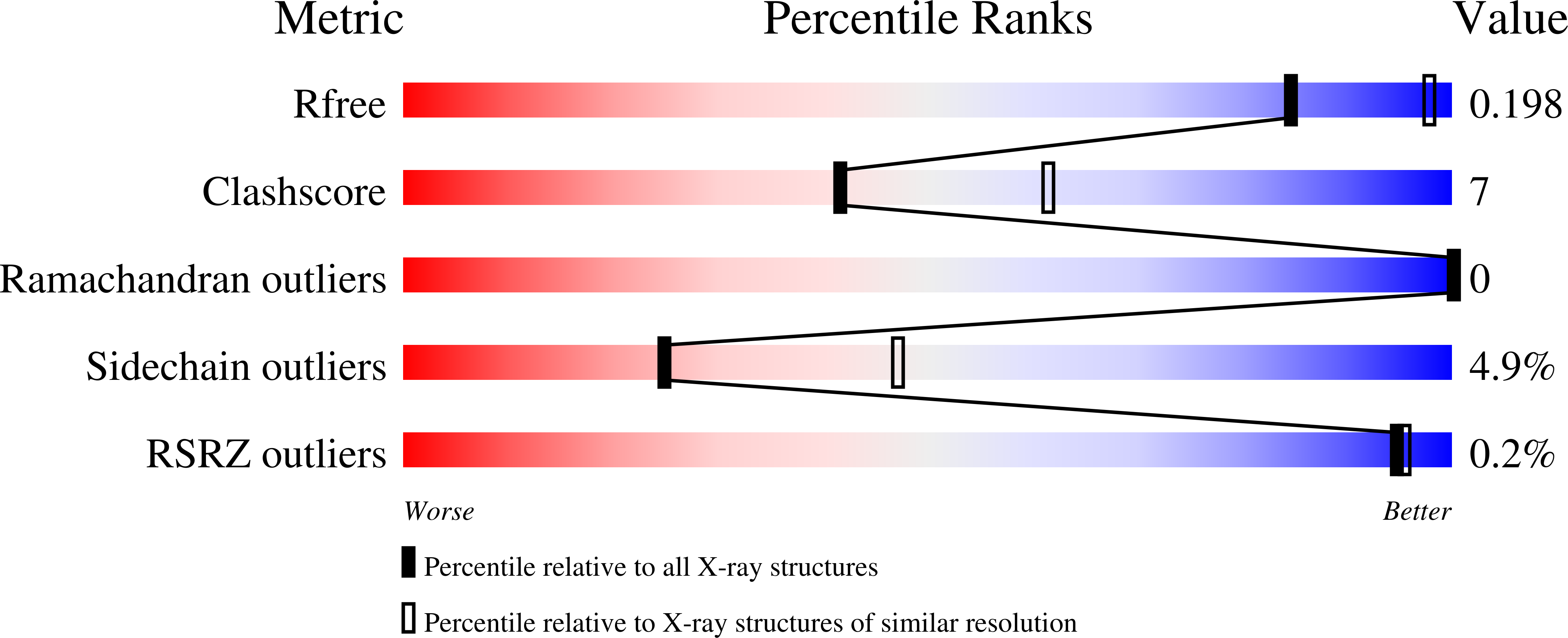
Switching between the Alternative Structures and Functions of a 2-Cys Peroxiredoxin, by Site-Directed Mutagenesis
Angelucci, F., Saccoccia, F., Ardini, M., Boumis, G., Brunori, M., Dileandro, L., Ippoliti, R., Miele, A.E., Natoli, G., Scotti, S., Bellelli, A.(2013) J Mol Biol 425: 4556
- PubMed: 24021815
- DOI: https://doi.org/10.1016/j.jmb.2013.09.002
- Primary Citation of Related Structures:
3ZL5, 3ZLP - PubMed Abstract:
Members of the typical 2-Cys peroxiredoxin (Prx) subfamily represent an intriguing example of protein moonlighting behavior since this enzyme shifts function: indeed, upon chemical stimuli, such as oxidative stress, Prx undergoes a switch from peroxidase to molecular chaperone, associated to a change in quaternary structure from dimers/decamers to higher-molecular-weight (HMW) species. In order to detail the structural mechanism of this switch at molecular level, we have designed and expressed mutants of peroxiredoxin I from Schistosoma mansoni (SmPrxI) with constitutive HMW assembly and molecular chaperone activity. By a combination of X-ray crystallography, transmission electron microscopy and functional experiments, we defined the structural events responsible for the moonlighting behavior of 2-Cys Prx and we demonstrated that acidification is coupled to local structural variations localized at the active site and a change in oligomerization to HMW forms, similar to those induced by oxidative stress. Moreover, we suggest that the binding site of the unfolded polypeptide is at least in part contributed by the hydrophobic surface exposed by the unfolding of the active site. We also find an inverse correlation between the extent of ring stacking and molecular chaperone activity that is explained assuming that the binding occurs at the extremities of the nanotube, and the longer the nanotube is, the lesser the ratio binding sites/molecular mass is.
Organizational Affiliation:
Department of Health, Life and Environmental Sciences, University of L'Aquila, Piazzale Salvatore Tommasi 1, 67100 L'Aquila, Italy. Electronic address: francesco.angelucci@univaq.it.