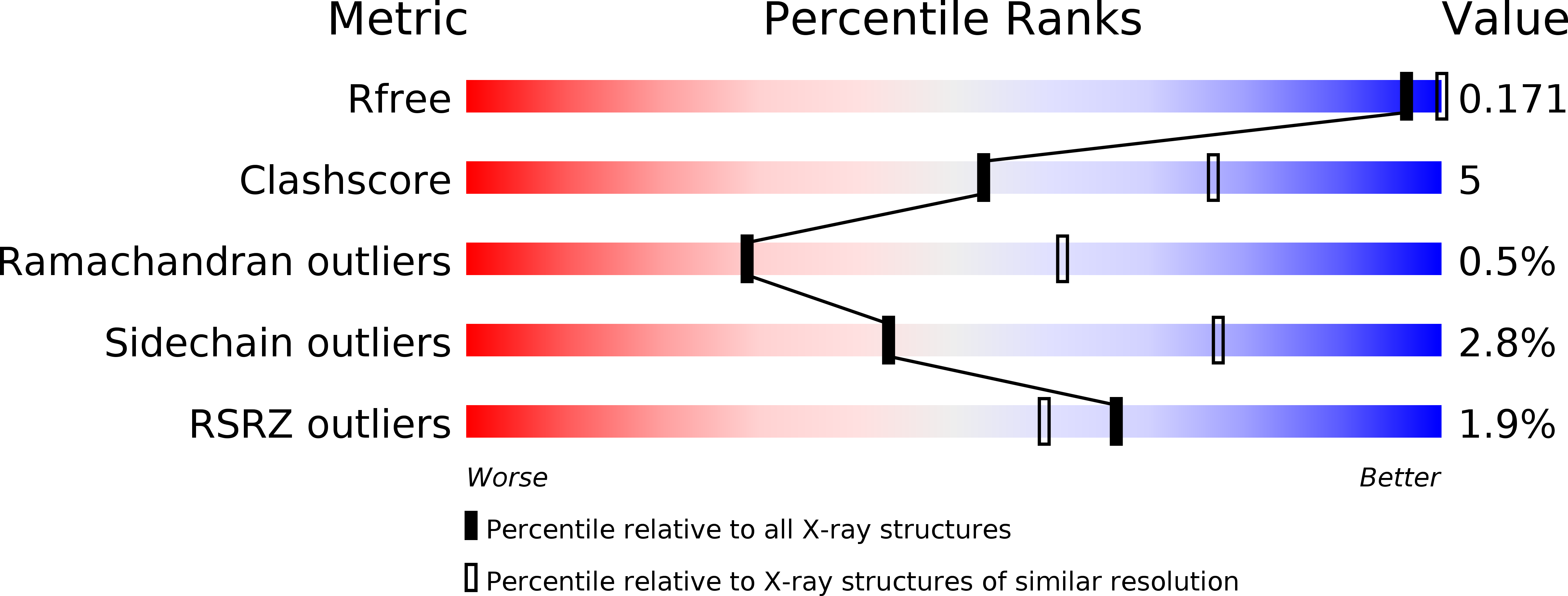
Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Arai, S., Yonezawa, Y., Okazaki, N., Matsumoto, F., Shibazaki, C., Shimizu, R., Yamada, M., Adachi, M., Tamada, T., Kawamoto, M., Tokunaga, H., Ishibashi, M., Blaber, M., Tokunaga, M., Kuroki, R.(2015) Acta Crystallogr D Biol Crystallogr 71: 541-554
- PubMed: 25760604
- DOI: https://doi.org/10.1107/S1399004714027734
- Primary Citation of Related Structures:
3WRT, 3WRZ, 3WS0, 3WS1, 3WS2, 3WS4, 3WS5 - PubMed Abstract:
Environmentally friendly absorbents are needed for Sr(2+) and Cs(+), as the removal of the radioactive Sr(2+) and Cs(+) that has leaked from the Fukushima Nuclear Power Plant is one of the most important problems in Japan. Halophilic proteins are known to have many acidic residues on their surface that can provide specific binding sites for metal ions such as Cs(+) or Sr(2+). The crystal structure of a halophilic β-lactamase from Chromohalobacter sp. 560 (HaBLA) was determined to resolutions of between 1.8 and 2.9 Å in space group P31 using X-ray crystallography. Moreover, the locations of bound Sr(2+) and Cs(+) ions were identified by anomalous X-ray diffraction. The location of one Cs(+)-specific binding site was identified in HaBLA even in the presence of a ninefold molar excess of Na(+) (90 mM Na(+)/10 mM Cs(+)). From an activity assay using isothermal titration calorimetry, the bound Sr(2+) and Cs(+) ions do not significantly affect the enzymatic function of HaBLA. The observation of a selective and high-affinity Cs(+)-binding site provides important information that is useful for the design of artificial Cs(+)-binding sites that may be useful in the bioremediation of radioactive isotopes.
Organizational Affiliation:
Quantum Beam Science Directorate, Japan Atomic Energy Agency, 2-4 Shirakata-shirane, Tokai, Ibaraki 319-1195, Japan.