The Gal3p transducer of the GAL regulon interacts with the Gal80p repressor in its ligand-induced closed conformation.
Lavy, T., Kumar, P.R., He, H., Joshua-Tor, L.(2012) Genes Dev 26: 294-303
- PubMed: 22302941
- DOI: https://doi.org/10.1101/gad.182691.111
- Primary Citation of Related Structures:
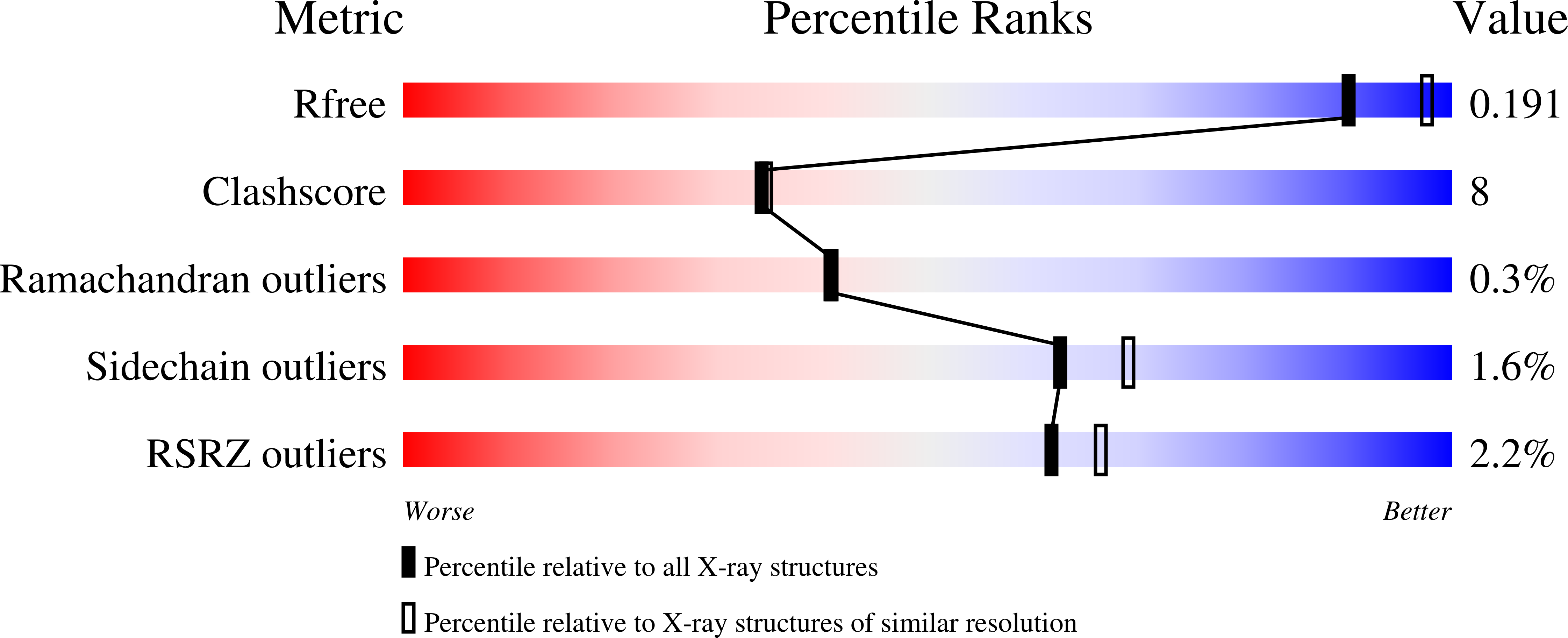
3V2U, 3V5R - PubMed Abstract:
A wealth of genetic information and some biochemical analysis have made the GAL regulon of the yeast Saccharomyces cerevisiae a classic model system for studying transcriptional activation in eukaryotes. Galactose induces this transcriptional switch, which is regulated by three proteins: the transcriptional activator Gal4p, bound to DNA; the repressor Gal80p; and the transducer Gal3p. We showed previously that NADP appears to act as a trigger to kick the repressor off the activator. Sustained activation involves a complex of the transducer Gal3p and Gal80p mediated by galactose and ATP. We solved the crystal structure of the complex of Gal3p-Gal80p with α-D-galactose and ATP to 2.1 Å resolution. The interaction between the proteins occurs only when Gal3p is in a "closed" state induced by ligand binding. The structure of the complex provides a rationale for the phenotypes of several well-known Gal80p and Gal3p mutants as well as the lack of galactokinase activity of Gal3p.
Organizational Affiliation:
Keck Structural Biology Laboratory, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 11724, USA.