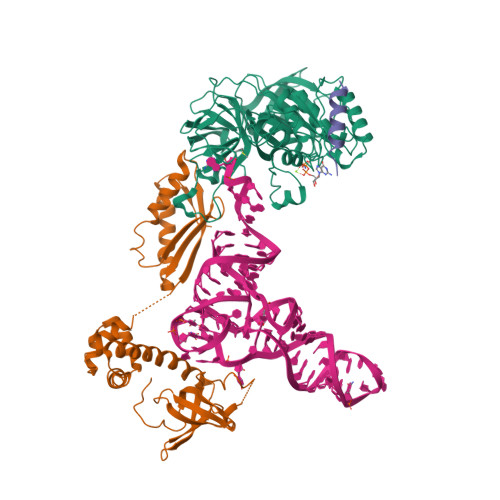
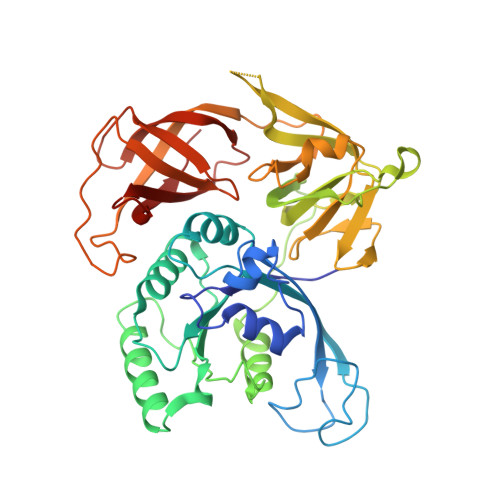
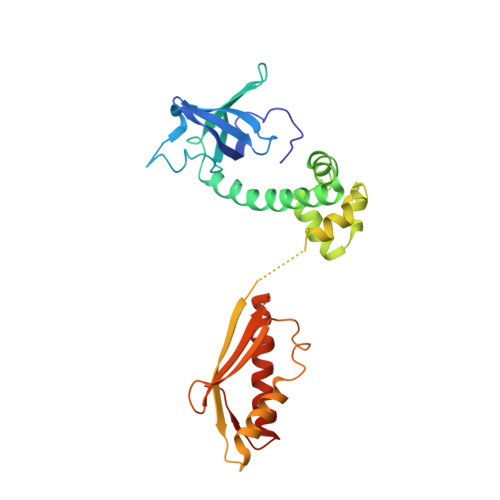
Structure of the ternary initiation complex aIF2-GDPNP-methionylated initiator tRNA.
Schmitt, E., Panvert, M., Lazennec-Schurdevin, C., Coureux, P.D., Perez, J., Thompson, A., Mechulam, Y.(2012) Nat Struct Mol Biol 19: 450-454
- PubMed: 22447243
- DOI: https://doi.org/10.1038/nsmb.2259
- Primary Citation of Related Structures:
3V11 - PubMed Abstract:
Eukaryotic and archaeal translation initiation factor 2 (e/aIF2) is a heterotrimeric GTPase that has a crucial role in the selection of the correct start codon on messenger RNA. We report the 5-Å resolution crystal structure of the ternary complex formed by archaeal aIF2 from Sulfolobus solfataricus, the GTP analog GDPNP and methionylated initiator tRNA. The 3D model is further supported by solution studies using small-angle X-ray scattering. The tRNA is bound by the α and γ subunits of aIF2. Contacts involve the elbow of the tRNA and the minor groove of the acceptor stem, but not the T-stem minor groove. We conclude that despite considerable structural homology between the core γ subunit of aIF2 and the elongation factor EF1A, these two G proteins of the translation apparatus use very different tRNA-binding strategies.
Organizational Affiliation:
Laboratoire de Biochimie, Unité mixte de Recherche 7654, Ecole Polytechnique, Centre National de la Recherche Scientifique, Palaiseau, France. emma@botrytis.polytechnique.fr