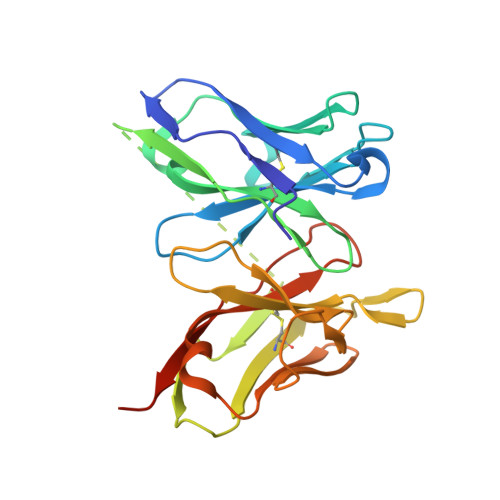
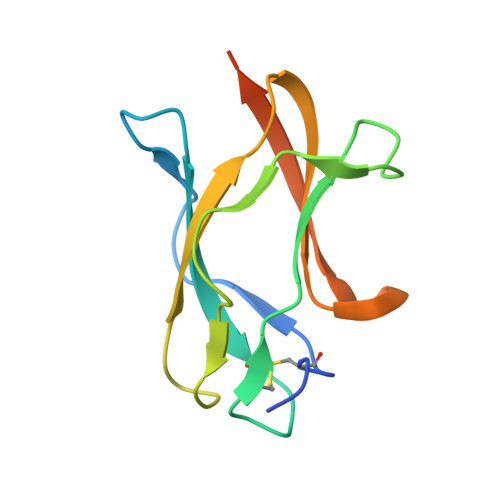
Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody.
Cockburn, J.J., Navarro Sanchez, M.E., Fretes, N., Urvoas, A., Staropoli, I., Kikuti, C.M., Coffey, L.L., Arenzana Seisdedos, F., Bedouelle, H., Rey, F.A.(2012) Structure 20: 303-314
- PubMed: 22285214
- DOI: https://doi.org/10.1016/j.str.2012.01.001
- Primary Citation of Related Structures:
3UYP, 3UZE, 3UZQ, 3UZV - PubMed Abstract:
The dengue virus (DENV) complex is composed of four distinct but serologically related flaviviruses, which together cause the present-day most important emerging viral disease. Although DENV infection induces lifelong immunity against viruses of the same serotype, the antibodies raised appear to contribute to severe disease in cases of heterotypic infections. Understanding the mechanisms of DENV neutralization by antibodies is, therefore, crucial for the design of vaccines that simultaneously protect against all four viruses. Here, we report a comparative, high-resolution crystallographic analysis of an "A-strand" murine monoclonal antibody, Mab 4E11, in complex with its target domain of the envelope protein from the four DENVs. Mab 4E11 is capable of neutralizing all four serotypes, and our study reveals the determinants of this cross-reactivity. The structures also highlight the mechanism by which A-strand Mabs disrupt the architecture of the mature virion, inducing premature fusion loop exposure and concomitant particle inactivation.
Organizational Affiliation:
Unité de Virologie Structurale, Département de Virologie, Institut Pasteur, 75724 Paris Cedex 15, France.