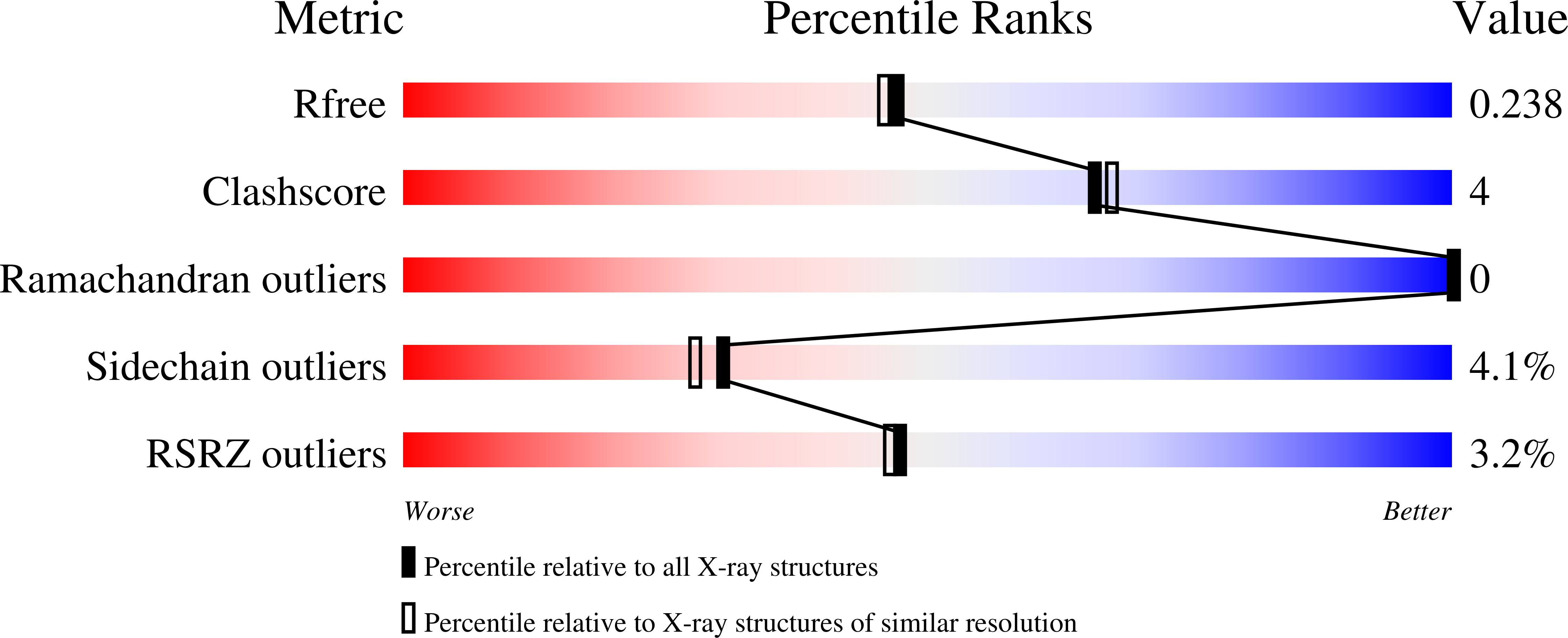
The role of the beta5-alpha11 loop in the active-site dynamics of acylated penicillin-binding protein A from Mycobacterium tuberculosis
Fedarovich, A., Nicholas, R.A., Davies, C.(2012) J Mol Biol 418: 316-330
- PubMed: 22365933
- DOI: https://doi.org/10.1016/j.jmb.2012.02.021
- Primary Citation of Related Structures:
3UN7, 3UPN, 3UPO, 3UPP - PubMed Abstract:
Penicillin-binding protein A (PBPA) is a class B penicillin-binding protein that is important for cell division in Mycobacterium tuberculosis. We have determined a second crystal structure of PBPA in apo form and compared it with an earlier structure of apoenzyme. Significant structural differences in the active site region are apparent, including increased ordering of a β-hairpin loop and a shift of the SxN active site motif such that it now occupies a position that appears catalytically competent. Using two assays, including one that uses the intrinsic fluorescence of a tryptophan residue, we have also measured the second-order acylation rate constants for the antibiotics imipenem, penicillin G, and ceftriaxone. Of these, imipenem, which has demonstrable anti-tubercular activity, shows the highest acylation efficiency. Crystal structures of PBPA in complex with the same antibiotics were also determined, and all show conformational differences in the β5-α11 loop near the active site, but these differ for each β-lactam and also for each of the two molecules in the crystallographic asymmetric unit. Overall, these data reveal the β5-α11 loop of PBPA as a flexible region that appears important for acylation and provide further evidence that penicillin-binding proteins in apo form can occupy different conformational states.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Medical University of South Carolina, Charleston, SC 29425, USA.