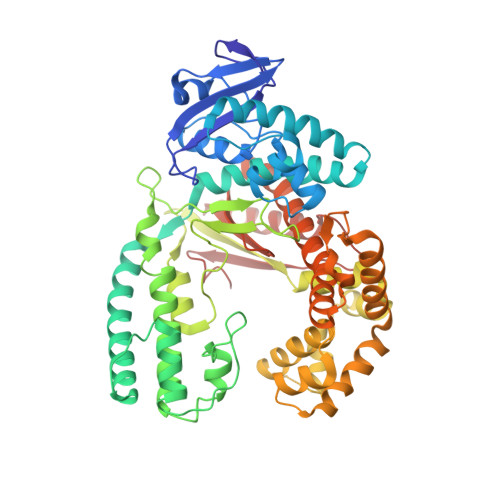
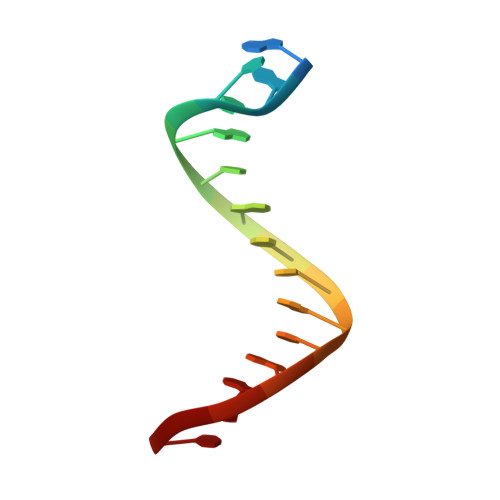
KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Betz, K., Malyshev, D.A., Lavergne, T., Welte, W., Diederichs, K., Dwyer, T.J., Ordoukhanian, P., Romesberg, F.E., Marx, A.(2012) Nat Chem Biol 8: 612-614
- PubMed: 22660438
- DOI: https://doi.org/10.1038/nchembio.966
- Primary Citation of Related Structures:
3RTV, 3SV3, 3SV4, 3SYZ, 3SZ2 - PubMed Abstract:
Many candidate unnatural DNA base pairs have been developed, but some of the best-replicated pairs adopt intercalated structures in free DNA that are difficult to reconcile with known mechanisms of polymerase recognition. Here we present crystal structures of KlenTaq DNA polymerase at different stages of replication for one such pair, dNaM-d5SICS, and show that efficient replication results from the polymerase itself, inducing the required natural-like structure.
Organizational Affiliation:
Department of Chemistry, Universität Konstanz, Konstanz, Germany.