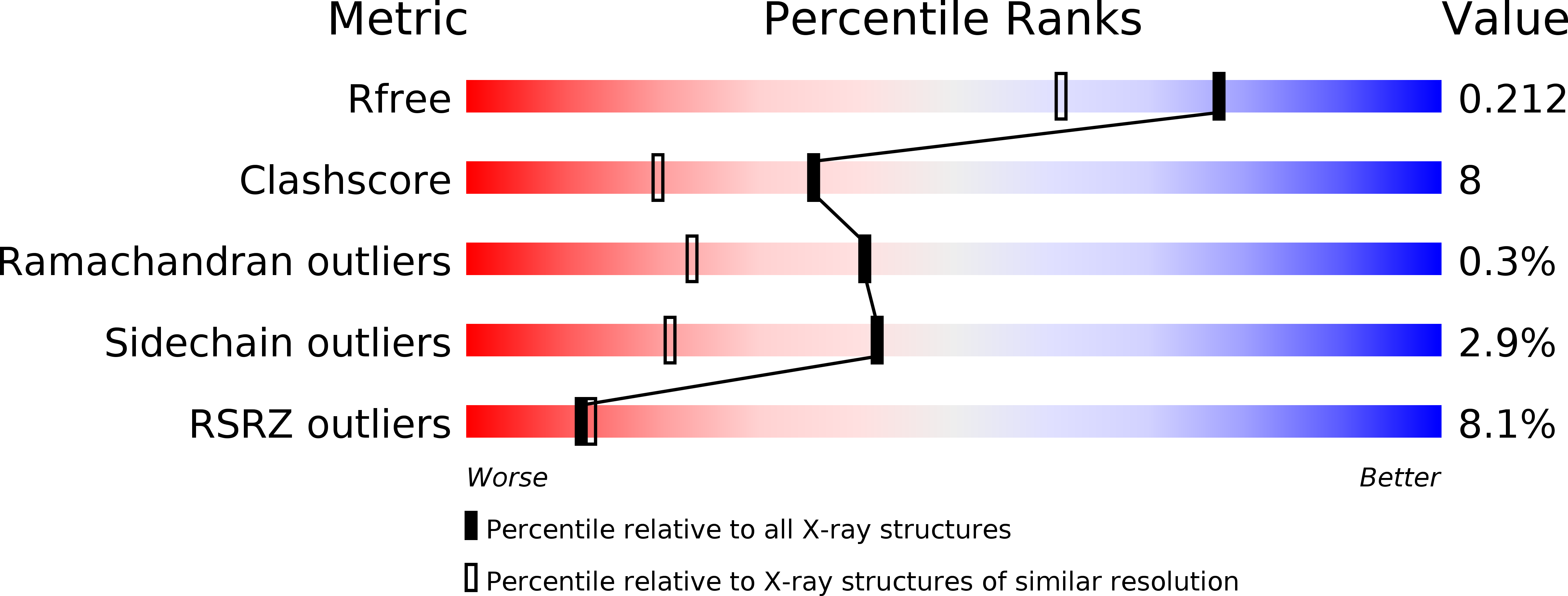
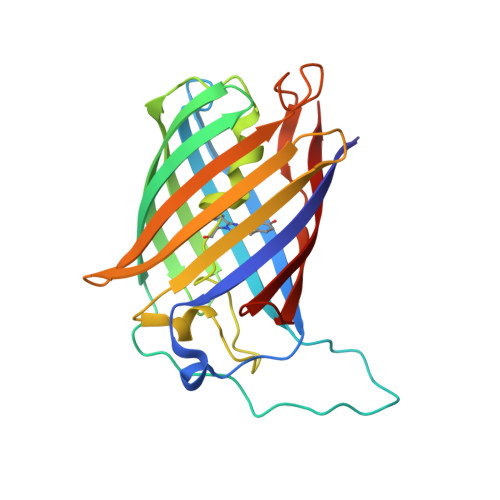
Circular permutation of red fluorescent proteins.
Shui, B., Wang, Q., Lee, F., Byrnes, L.J., Chudakov, D.M., Lukyanov, S.A., Sondermann, H., Kotlikoff, M.I.(2011) PLoS One 6: e20505-e20505
- PubMed: 21647365
- DOI: https://doi.org/10.1371/journal.pone.0020505
- Primary Citation of Related Structures:
3RWA, 3RWT - PubMed Abstract:
Circular permutation of fluorescent proteins provides a substrate for the design of molecular sensors. Here we describe a systematic exploration of permutation sites for mCherry and mKate using a tandem fusion template approach. Circular permutants retaining more than 60% (mCherry) and 90% (mKate) brightness of the parent molecules are reported, as well as a quantitative evaluation of the fluorescence from neighboring mutations. Truncations of circular permutants indicated essential N- and C-terminal segments and substantial flexibility in the use of these molecules. Structural evaluation of two cp-mKate variants indicated no major conformational changes from the previously reported wild-type structure, and cis conformation of the chromophores. Four cp-mKates were identified with over 80% of native fluorescence, providing important new building blocks for sensor and complementation experiments.
Organizational Affiliation:
Department of Biomedical Sciences, College of Veterinary Medicine, Cornell University, Ithaca, New York, United States of America.