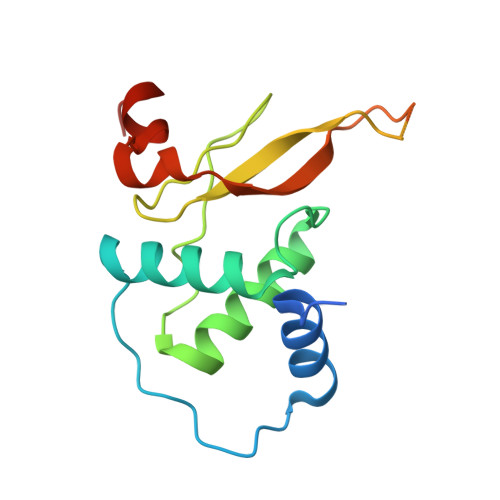
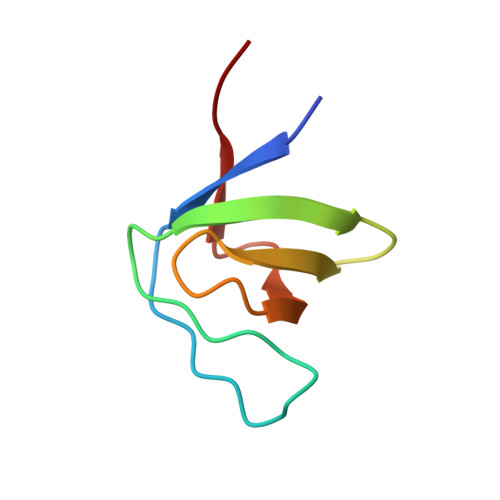
Molecular design, functional characterization and structural basis of a protein inhibitor against the HIV-1 pathogenicity factor Nef.
Breuer, S., Schievink, S.I., Schulte, A., Blankenfeldt, W., Fackler, O.T., Geyer, M.(2011) PLoS One 6: e20033-e20033
- PubMed: 21625496
- DOI: https://doi.org/10.1371/journal.pone.0020033
- Primary Citation of Related Structures:
3REA, 3REB - PubMed Abstract:
Increased spread of HIV-1 and rapid emergence of drug resistance warrants development of novel antiviral strategies. Nef, a critical viral pathogenicity factor that interacts with host cell factors but lacks enzymatic activity, is not targeted by current antiviral measures. Here we inhibit Nef function by simultaneously blocking several highly conserved protein interaction surfaces. This strategy, referred to as "wrapping Nef", is based on structure-function analyses that led to the identification of four target sites: (i) SH3 domain interaction, (ii) interference with protein transport processes, (iii) CD4 binding and (iv) targeting to lipid membranes. Screening combinations of Nef-interacting domains, we developed a series of small Nef interacting proteins (NIs) composed of an SH3 domain optimized for binding to Nef, fused to a sequence motif of the CD4 cytoplasmic tail and combined with a prenylation signal for membrane association. NIs bind to Nef in the low nM affinity range, associate with Nef in human cells and specifically interfere with key biological activities of Nef. Structure determination of the Nef-inhibitor complex reveals the molecular basis for binding specificity. These results establish Nef-NI interfaces as promising leads for the development of potent Nef inhibitors.
Organizational Affiliation:
Abteilung Physikalische Biochemie, Max-Planck-Institut für molekulare Physiologie, Dortmund, Germany.