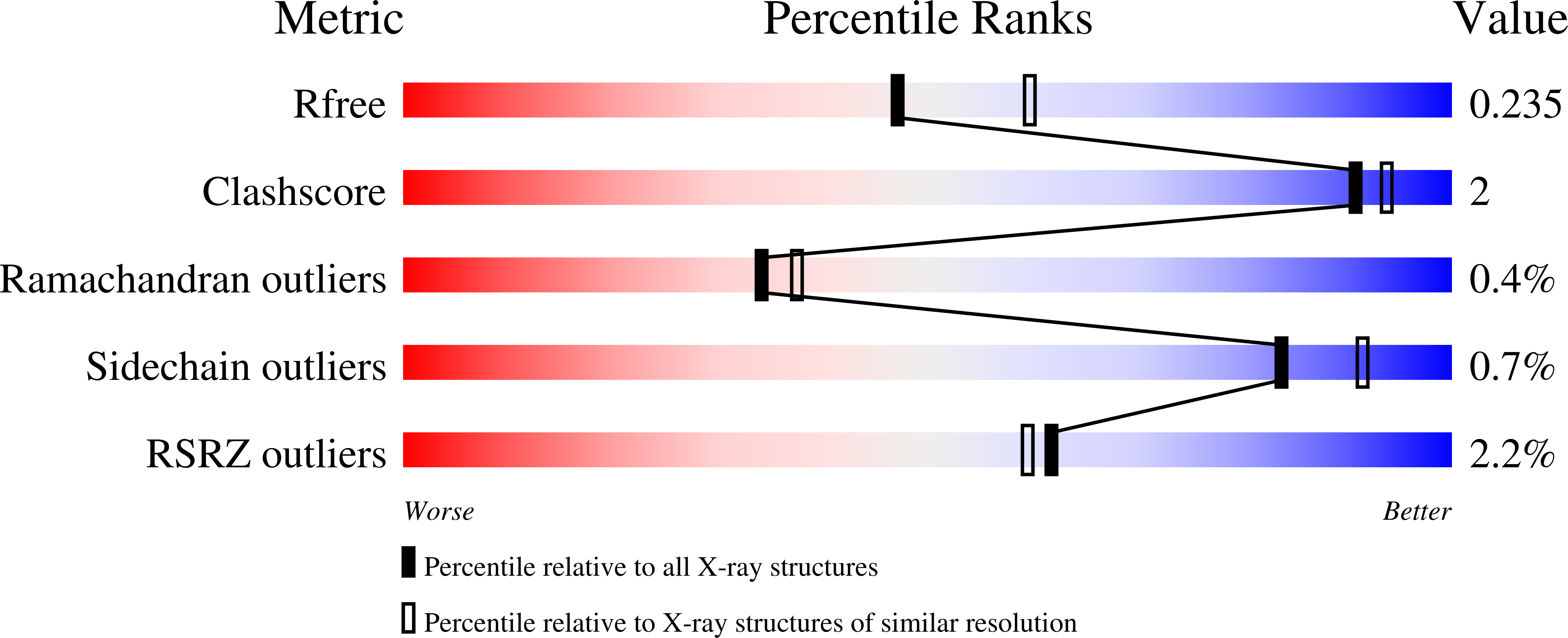
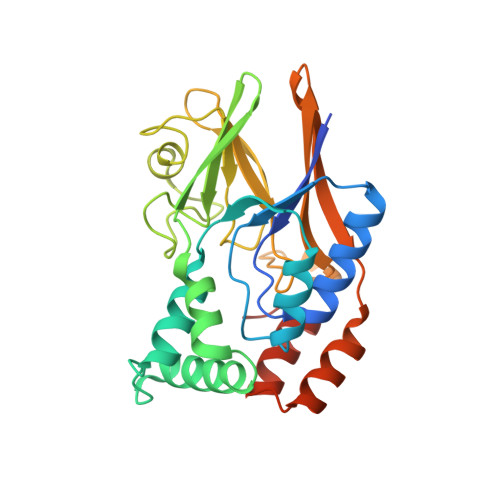
Structural and enzymatic characterization of a Streptococcal ATP/diadenosine polyphosphate and phosphodiester hydrolase Spr1479/SapH
Jiang, Y.L., Zhang, J.W., Yu, W.L., Cheng, W., Zhang, C.C., Frolet, C., Di-guilmi, A.-M., Vernet, T., Zhou, C.Z., Chen, Y.To be published.