Structural basis for inhibition of bacterial ATP synthase by subunit epsilon of the rotor stalk
Cingolani, G., Duncan, T.M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP synthase subunit alpha | 513 | Escherichia coli DH1 | Mutation(s): 0 Gene Names: atpA, EcDH1_4233 EC: 3.6.3.14 Membrane Entity: Yes | 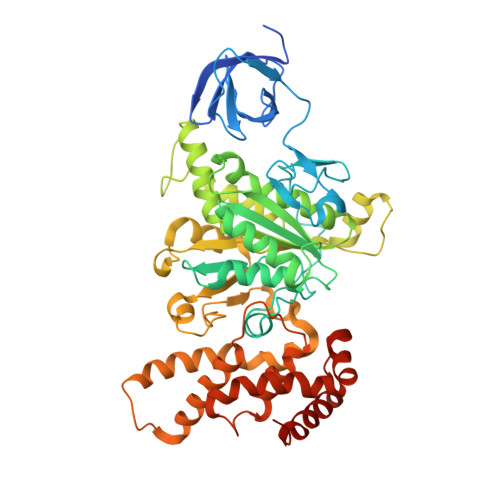 | |
UniProt | |||||
Find proteins for P0ABB0 (Escherichia coli (strain K12)) Explore P0ABB0 Go to UniProtKB: P0ABB0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0ABB0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP synthase subunit beta | 459 | Escherichia coli DH1 | Mutation(s): 1 Gene Names: atpD, EcDH1_4235 EC: 3.6.3.14 Membrane Entity: Yes | 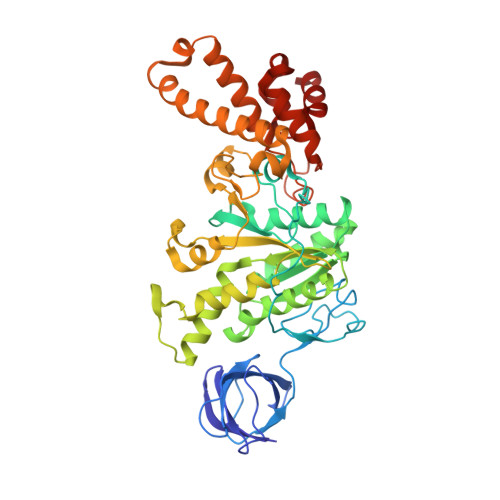 | |
UniProt | |||||
Find proteins for P0ABB4 (Escherichia coli (strain K12)) Explore P0ABB4 Go to UniProtKB: P0ABB4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0ABB4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP synthase gamma chain | EA [auth e], G, O, W | 286 | Escherichia coli DH1 | Mutation(s): 0 Gene Names: EcDH1_4234 Membrane Entity: Yes |  |
UniProt | |||||
Find proteins for P0ABA6 (Escherichia coli (strain K12)) Explore P0ABA6 Go to UniProtKB: P0ABA6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0ABA6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP synthase epsilon chain | FA [auth f], H, P, X | 138 | Escherichia coli DH1 | Mutation(s): 0 Gene Names: atpC, EcDH1_4236 Membrane Entity: Yes | 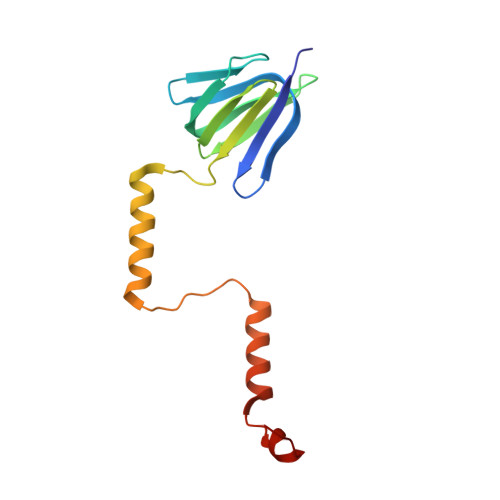 |
UniProt | |||||
Find proteins for P0A6E6 (Escherichia coli (strain K12)) Explore P0A6E6 Go to UniProtKB: P0A6E6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A6E6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ANP Query on ANP | GA [auth A] GB [auth Q] IA [auth B] IB [auth R] KA [auth C] | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER C10 H17 N6 O12 P3 PVKSNHVPLWYQGJ-KQYNXXCUSA-N |  | ||
| ADP Query on ADP | MA [auth D], MB [auth T], YB [auth b], ZA [auth L] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| SO4 Query on SO4 | AC [auth b] BB [auth L] BC [auth c] CB [auth M] CC [auth d] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| MG Query on MG | AB [auth L] HA [auth A] HB [auth Q] JA [auth B] JB [auth R] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 435.97 | α = 90 |
| b = 183 | β = 108.99 |
| c = 225.39 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data collection |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |