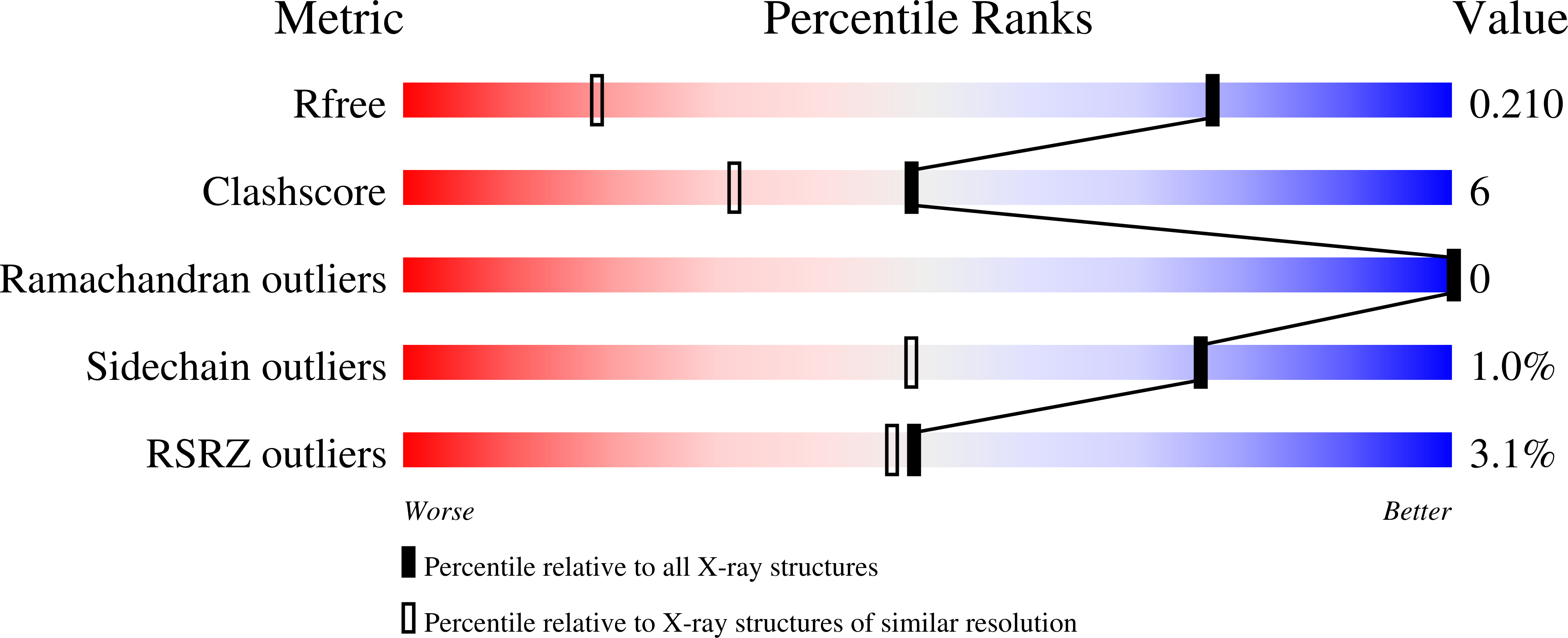
Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90.
Bracher, A., Kozany, C., Thost, A.K., Hausch, F.(2011) Acta Crystallogr D Biol Crystallogr 67: 549-559
- PubMed: 21636895
- DOI: https://doi.org/10.1107/S0907444911013862
- Primary Citation of Related Structures:
3O5D, 3O5E, 3O5F, 3O5G, 3O5I, 3O5J, 3O5K, 3O5L, 3O5M, 3O5O, 3O5P, 3O5Q, 3O5R - PubMed Abstract:
Steroid hormone receptors are key components of mammalian stress and sex hormone systems. Many of them rely on the Hsp90 chaperone system for full function and are further fine-tuned by Hsp90-associated peptidyl-prolyl isomerases such as FK506-binding proteins 51 and 52. FK506-binding protein 51 (FKBP51) has been shown to reduce glucocorticoid receptor signalling and has been genetically associated with human stress resilience and with numerous psychiatric disorders. The peptidyl-prolyl isomerase domain of FKBP51 contains a high-affinity binding site for the natural products FK506 and rapamycin and has further been shown to convey most of the inhibitory activity on the glucocorticoid receptor. FKBP51 has therefore become a prime new target for the treatment of stress-related affective disorders that could be amenable to structure-based drug design. Here, a series of high-resolution structures of the peptidyl-prolyl isomerase domain of FKBP51 as well as a cocrystal structure with the prototypic ligand FK506 are described. These structures provide a detailed picture of the drug-binding domain of FKBP51 and the molecular binding mode of its ligand as a starting point for the rational design of improved inhibitors.
Organizational Affiliation:
Department of Cellular Biochemistry, Max Planck Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried, Germany. bracher@biochem.mpg.de