Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
Osipiuk, J., Mulligan, R., Bargassa, M., Hamilton, J.E., Cunningham, M.A., Joachimiak, A.(2012) J Biol Chem 287: 19452-19461
- PubMed: 22493430
- DOI: https://doi.org/10.1074/jbc.M112.358069
- Primary Citation of Related Structures:
3N55, 3NJF, 3NJG, 3NJH, 3NJI, 3NJJ, 3NJK, 3NJL, 3NJM, 3NJN - PubMed Abstract:
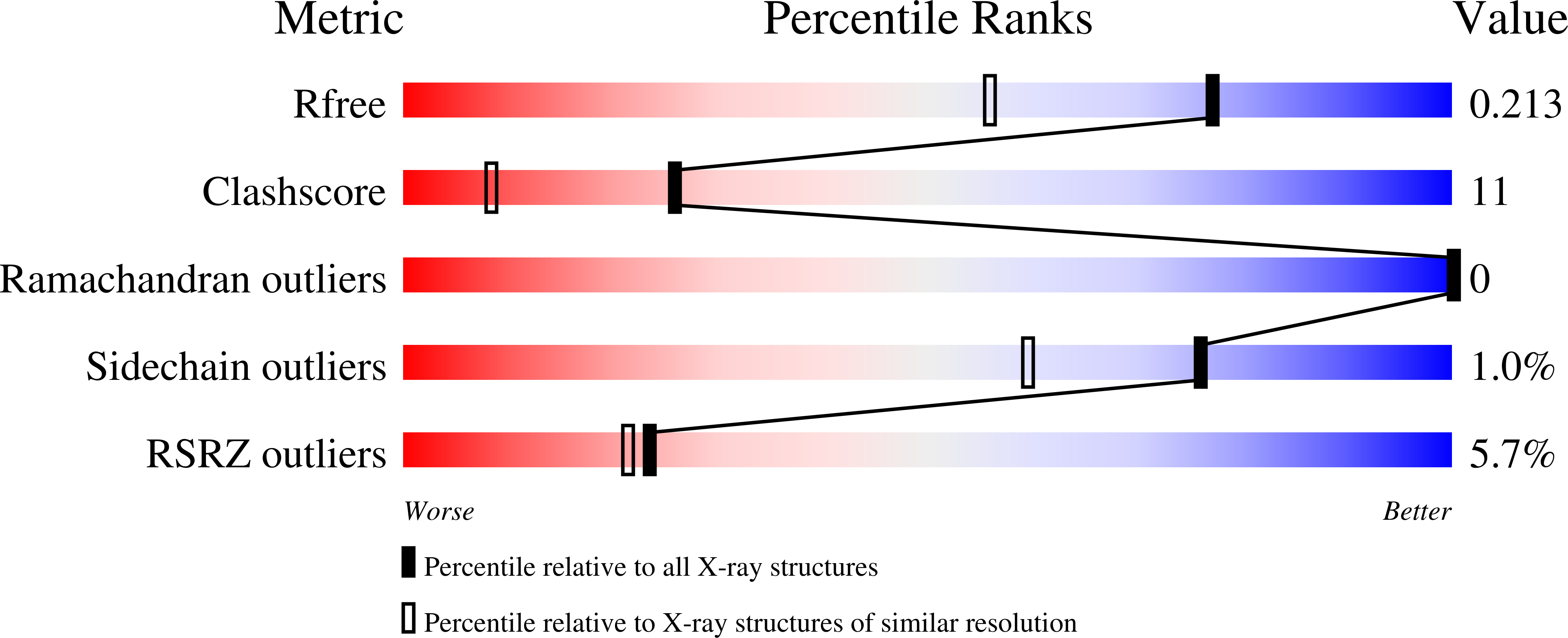
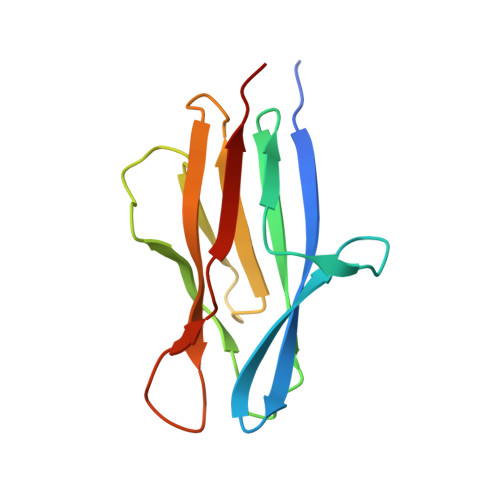
The crystal structure of SO1698 protein from Shewanella oneidensis was determined by a SAD method and refined to 1.57 Å. The structure is a β sandwich that unexpectedly consists of two polypeptides; the N-terminal fragment includes residues 1-116, and the C-terminal one includes residues 117-125. Electron density also displayed the Lys-98 side chain covalently linked to Asp-116. The putative active site residues involved in self-cleavage were identified; point mutants were produced and characterized structurally and in a biochemical assay. Numerical simulations utilizing molecular dynamics and hybrid quantum/classical calculations suggest a mechanism involving activation of a water molecule coordinated by a catalytic aspartic acid.
Organizational Affiliation:
Midwest Center for Structural Genomics and Structural Biology Center, Biosciences, Argonne National Laboratory, Argonne, Illinois 60439, USA.