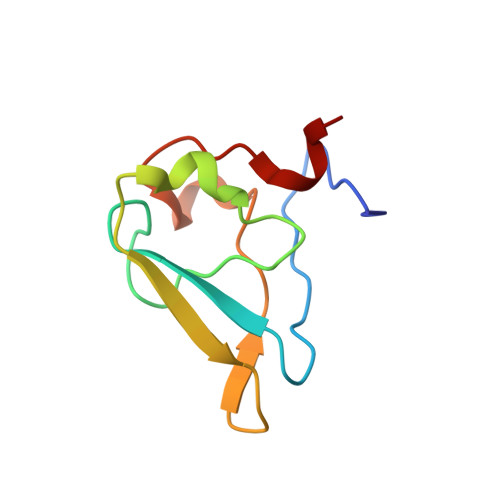
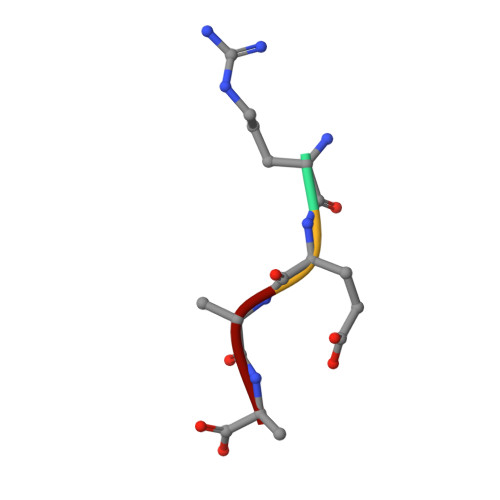
Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Choi, W.S., Jeong, B.-C., Joo, Y.J., Lee, M.-R., Kim, J., Eck, M.J., Song, H.K.(2010) Nat Struct Mol Biol 17: 1175-1181
- PubMed: 20835240
- DOI: https://doi.org/10.1038/nsmb.1907
- Primary Citation of Related Structures:
3NIH, 3NII, 3NIJ, 3NIK, 3NIL, 3NIM, 3NIN, 3NIS, 3NIT - PubMed Abstract:
The N-end rule pathway is a regulated proteolytic system that targets proteins containing destabilizing N-terminal residues (N-degrons) for ubiquitination and proteasomal degradation in eukaryotes. The N-degrons of type 1 substrates contain an N-terminal basic residue that is recognized by the UBR box domain of the E3 ubiquitin ligase UBR1. We describe structures of the UBR box of Saccharomyces cerevisiae UBR1 alone and in complex with N-degron peptides, including that of the cohesin subunit Scc1, which is cleaved and targeted for degradation at the metaphase-anaphase transition. The structures reveal a previously unknown protein fold that is stabilized by a novel binuclear zinc center. N-terminal arginine, lysine or histidine side chains of the N-degron are coordinated in a multispecific binding pocket. Unexpectedly, the structures together with our in vitro biochemical and in vivo pulse-chase analyses reveal a previously unknown modulation of binding specificity by the residue at position 2 of the N-degron.
Organizational Affiliation:
School of Life Sciences and Biotechnology, Korea University, Anam-Dong, Korea.