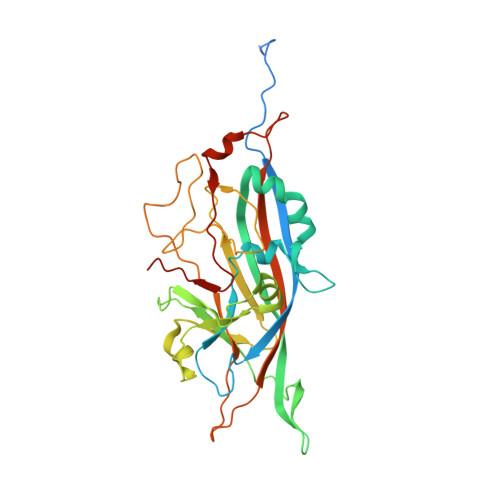
Structure of Penaeus stylirostris densovirus, a shrimp pathogen.
Kaufmann, B., Bowman, V.D., Li, Y., Szelei, J., Waddell, P.J., Tijssen, P., Rossmann, M.G.(2010) J Virol 84: 11289-11296
- PubMed: 20702621
- DOI: https://doi.org/10.1128/JVI.01240-10
- Primary Citation of Related Structures:
3N7X - PubMed Abstract:
Penaeus stylirostris densovirus (PstDNV), a pathogen of penaeid shrimp, causes significant damage to farmed and wild shrimp populations. In contrast to other parvoviruses, PstDNV probably has only one type of capsid protein that lacks the phospholipase A2 activity that has been implicated as a requirement during parvoviral host cell infection. The structure of recombinant virus-like particles, composed of 60 copies of the 37.5-kDa coat protein, the smallest parvoviral capsid protein reported thus far, was determined to 2.5-Å resolution by X-ray crystallography. The structure represents the first near-atomic resolution structure within the genus Brevidensovirus. The capsid protein has a β-barrel "jelly roll" motif similar to that found in many icosahedral viruses, including other parvoviruses. The N-terminal portion of the PstDNV coat protein adopts a "domain-swapped" conformation relative to its twofold-related neighbor similar to the insect parvovirus Galleria mellonella densovirus (GmDNV) but in stark contrast to vertebrate parvoviruses. However, most of the surface loops have little structural resemblance to any of the known parvoviral capsid proteins.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, 915 West State Street, West Lafayette, IN 47907-2054, USA.