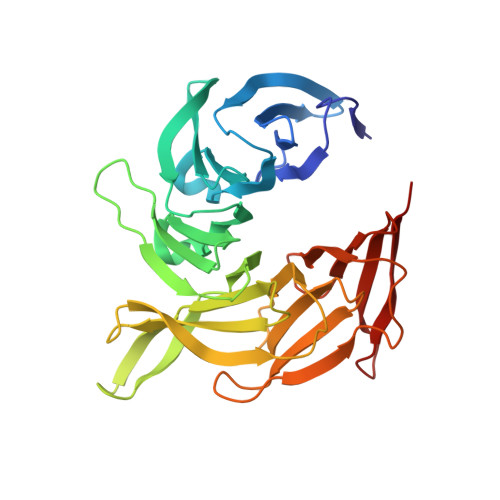
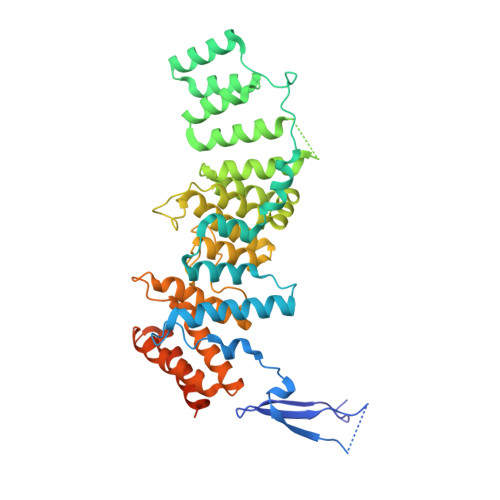
Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat.
Whittle, J.R., Schwartz, T.U.(2010) J Cell Biol 190: 347-361
- PubMed: 20696705
- DOI: https://doi.org/10.1083/jcb.201003092
- Primary Citation of Related Structures:
3MZK, 3MZL - PubMed Abstract:
Ancestral coatomer element 1 (ACE1) proteins assemble latticework coats for COPII vesicles and the nuclear pore complex. The ACE1 protein Sec31 and Sec13 make a 2:2 tetramer that forms the edge element of the COPII outer coat. In this study, we report that the COPII accessory protein Sec16 also contains an ACE1. The 165-kD crystal structure of the central domain of Sec16 in complex with Sec13 was solved at 2.7-A resolution. Sec16 and Sec13 also make a 2:2 tetramer, another edge element for the COPII system. Domain swapping at the ACE1-ACE1 interface is observed both in the prior structure of Sec13-Sec31 and in Sec13-Sec16. A Sec31 mutant in which domain swapping is prevented adopts an unprecedented laminated structure, solved at 2.8-A resolution. Our in vivo data suggest that the ACE1 element of Sec31 can functionally replace the ACE1 element of Sec16. Our data support Sec16 as a scaffold for the COPII system and a template for the Sec13-Sec31 coat.
Organizational Affiliation:
Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.