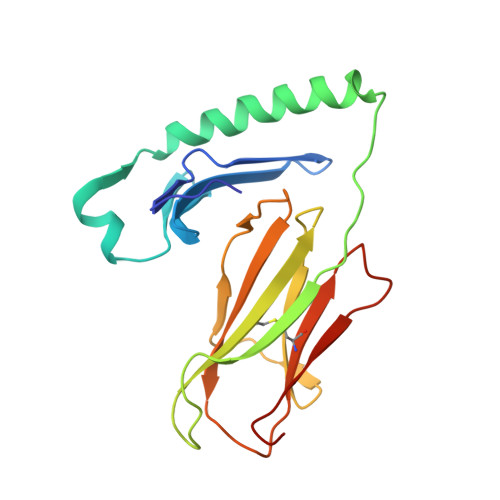
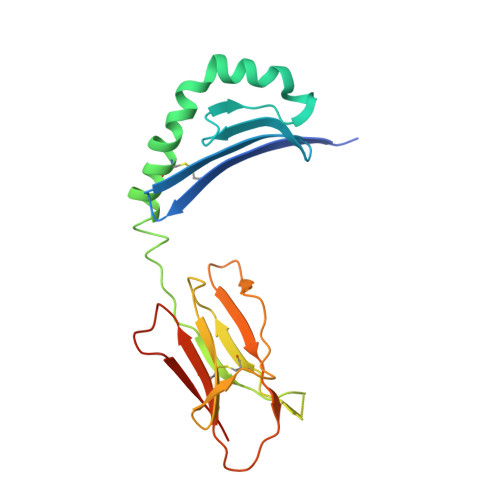
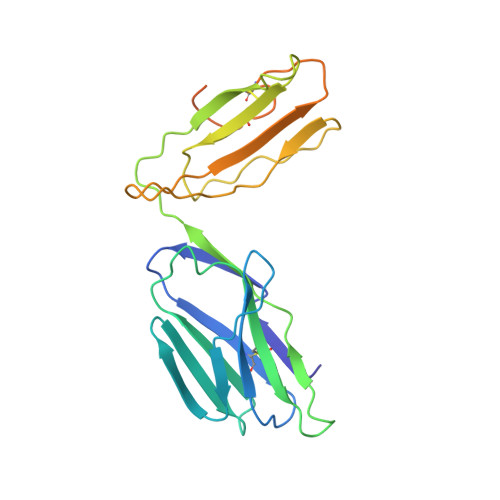
The diabetogenic mouse MHC class II molecule I-Ag7 is endowed with a switch that modulates TCR affinity.
Yoshida, K., Corper, A.L., Herro, R., Jabri, B., Wilson, I.A., Teyton, L.(2010) J Clin Invest 120: 1578-1590
- PubMed: 20407212
- DOI: https://doi.org/10.1172/JCI41502
- Primary Citation of Related Structures:
3MBE - PubMed Abstract:
Genetic susceptibility to autoimmunity is frequently associated with specific MHC alleles. Diabetogenic MHC class II molecules, such as human HLA-DQ8 and mouse I-Ag7, typically have a small, uncharged amino acid residue at position 57 of their beta chain (beta57); this results in the absence of a salt bridge between beta57 and Argalpha76, which is adjacent to the P9 pocket of the peptide-binding groove. However, the influence of Argalpha76 on the selection of the TCR repertoire remains unknown, particularly when the MHC molecule binds a peptide with a neutral amino acid residue at position P9. Here, we have shown that diabetogenic MHC class II molecules bound to a peptide with a neutral P9 residue primarily selected and expanded cells expressing TCRs bearing a negatively charged residue in the first segment of their complementarity determining region 3beta. The crystal structure of one such TCR in complex with I-Ag7 bound to a peptide containing a neutral P9 residue revealed that a network of favorable long-range (greater than 4 A) electrostatic interactions existed among Argalpha76, the neutral P9 residue, and TCR, which supported the substantially increased TCR/peptide-MHC affinity. This network could be modulated or switched to a lower affinity interaction by the introduction of a negative charge at position P9 of the peptide. Our results support the existence of a switch at residue beta57 of the I-Ag7 and HLA-DQ8 class II molecules and potentially link normal thymic TCR selection with abnormal peripheral behavior.
Organizational Affiliation:
Department of Immunology and Microbial Science, The Scripps Research Institute, La Jolla, California 92037, USA.