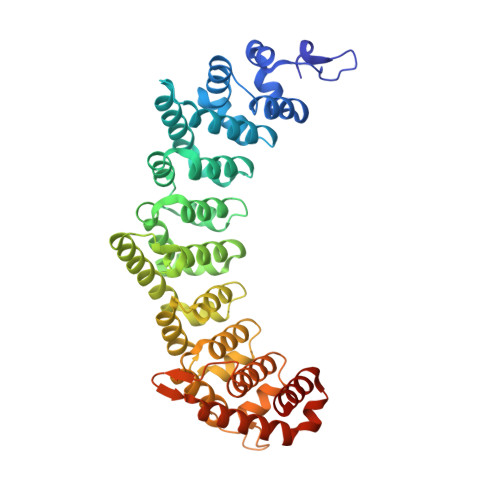
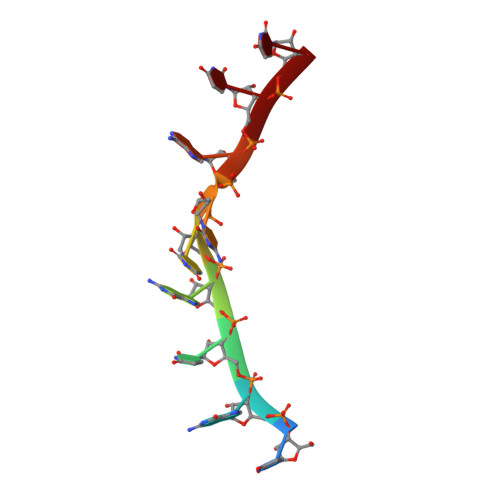
Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein.
Wang, Y., Opperman, L., Wickens, M., Hall, T.M.(2009) Proc Natl Acad Sci U S A 106: 20186-20191
- PubMed: 19901328
- DOI: https://doi.org/10.1073/pnas.0812076106
- Primary Citation of Related Structures:
3K5Q, 3K5Y, 3K5Z, 3K61, 3K62, 3K64 - PubMed Abstract:
Caenorhabditis elegans fem-3 binding factor (FBF) is a founding member of the PUMILIO/FBF (PUF) family of mRNA regulatory proteins. It regulates multiple mRNAs critical for stem cell maintenance and germline development. Here, we report crystal structures of FBF in complex with 6 different 9-nt RNA sequences, including elements from 4 natural mRNAs. These structures reveal that FBF binds to conserved bases at positions 1-3 and 7-8. The key specificity determinant of FBF vs. other PUF proteins lies in positions 4-6. In FBF/RNA complexes, these bases stack directly with one another and turn away from the RNA-binding surface. A short region of FBF is sufficient to impart its unique specificity and lies directly opposite the flipped bases. We suggest that this region imposes a flattened curvature on the protein; hence, the requirement for the additional nucleotide. The principles of FBF/RNA recognition suggest a general mechanism by which PUF proteins recognize distinct families of RNAs yet exploit very nearly identical atomic contacts in doing so.
Organizational Affiliation:
Laboratory of Structural Biology, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.