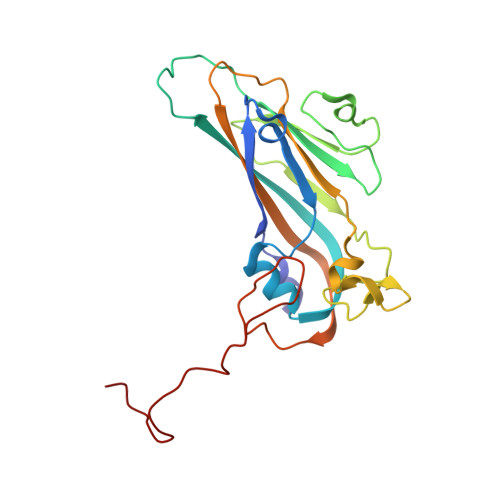
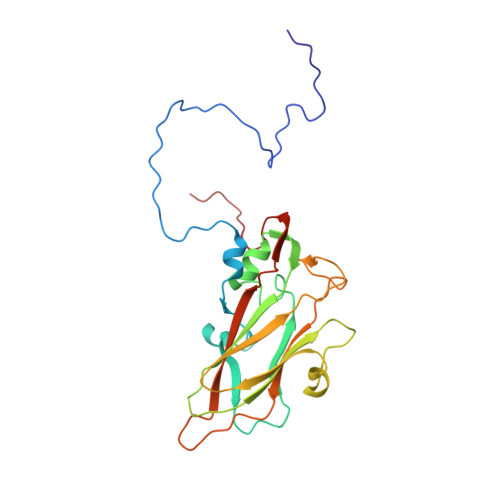
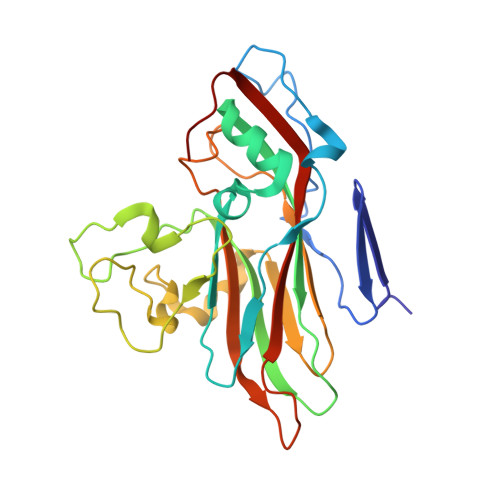
Structural and functional analysis of coxsackievirus A9 integrin {alpha}v{beta}6 binding and uncoating.
Shakeel, S., Seitsonen, J.J., Kajander, T., Laurinmaki, P., Hyypia, T., Susi, P., Butcher, S.J.(2013) J Virol 87: 3943-3951
- PubMed: 23365426
- DOI: https://doi.org/10.1128/JVI.02989-12
- Primary Citation of Related Structures:
3J2J - PubMed Abstract:
Coxsackievirus A9 (CVA9) is an important pathogen of the Picornaviridae family. It utilizes cellular receptors from the integrin αv family for binding to its host cells prior to entry and genome release. Among the integrins tested, it has the highest affinity for αvβ6, which recognizes the arginine-glycine-aspartic acid (RGD) loop present on the C terminus of viral capsid protein, VP1. As the atomic model of CVA9 lacks the RGD loop, we used surface plasmon resonance, electron cryo-microscopy, and image reconstruction to characterize the capsid-integrin interactions and the conformational changes on genome release. We show that the integrin binds to the capsid with nanomolar affinity and that the binding of integrin to the virion does not induce uncoating, thereby implying that further steps are required for release of the genome. Electron cryo-tomography and single-particle image reconstruction revealed variation in the number and conformation of the integrins bound to the capsid, with the integrin footprint mapping close to the predicted site for the exposed RGD loop on VP1. Comparison of empty and RNA-filled capsid reconstructions showed that the capsid undergoes conformational changes when the genome is released, so that the RNA-capsid interactions in the N termini of VP1 and VP4 are lost, VP4 is removed, and the capsid becomes more porous, as has been reported for poliovirus 1, human rhinovirus 2, enterovirus 71, and coxsackievirus A7. These results are important for understanding the structural basis of integrin binding to CVA9 and the molecular events leading to CVA9 cell entry and uncoating.
Organizational Affiliation:
Institute of Biotechnology, University of Helsinki, Helsinki, Finland.