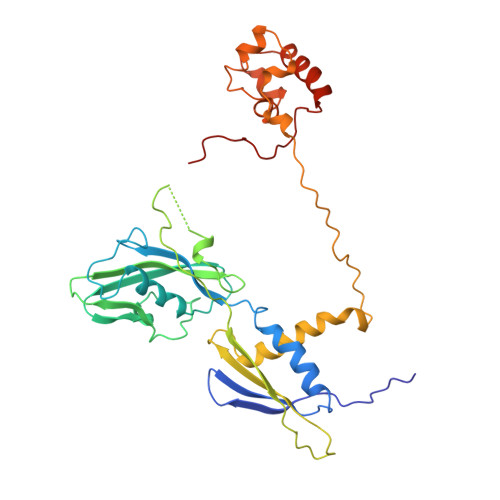
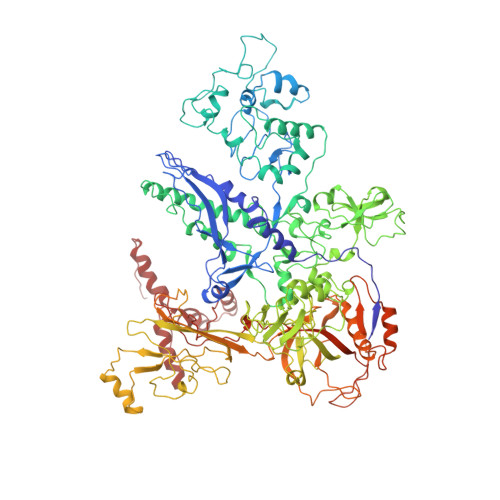
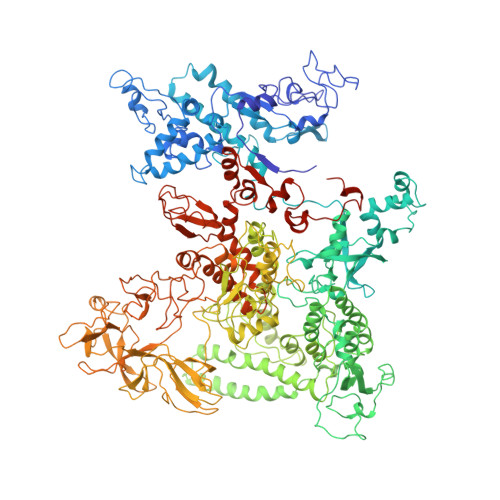
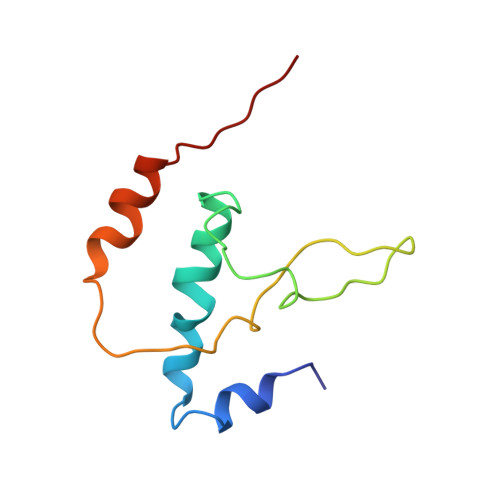
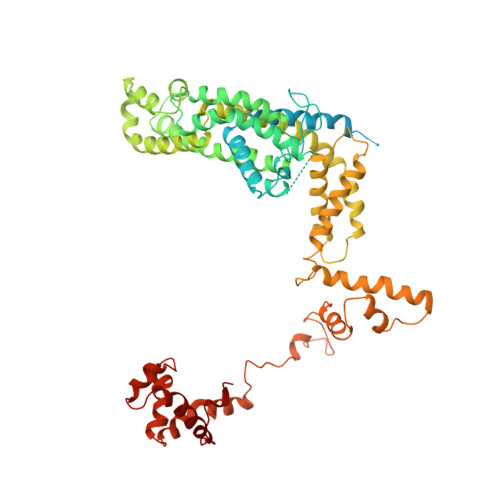
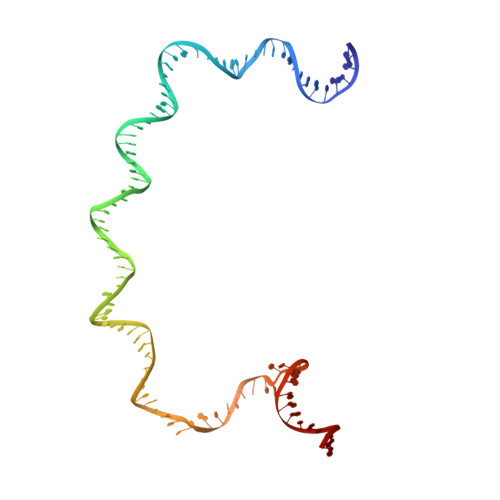
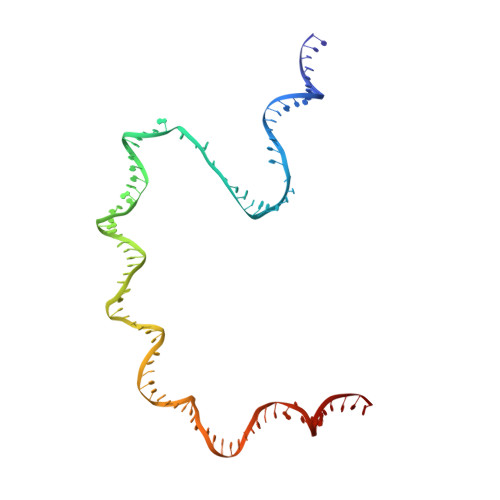
Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Hudson, B.P., Quispe, J., Lara-Gonzalez, S., Kim, Y., Berman, H.M., Arnold, E., Ebright, R.H., Lawson, C.L.(2009) Proc Natl Acad Sci U S A 106: 19830-19835
- PubMed: 19903881
- DOI: https://doi.org/10.1073/pnas.0908782106
- Primary Citation of Related Structures:
3IYD - PubMed Abstract:
We present the experimentally determined 3D structure of an intact activator-dependent transcription initiation complex comprising the Escherichia coli catabolite activator protein (CAP), RNA polymerase holoenzyme (RNAP), and a DNA fragment containing positions -78 to +20 of a Class I CAP-dependent promoter with a CAP site at position -61.5 and a premelted transcription bubble. A 20-A electron microscopy reconstruction was obtained by iterative projection-based matching of single particles visualized in carbon-sandwich negative stain and was fitted using atomic coordinate sets for CAP, RNAP, and DNA. The structure defines the organization of a Class I CAP-RNAP-promoter complex and supports previously proposed interactions of CAP with RNAP alpha subunit C-terminal domain (alphaCTD), interactions of alphaCTD with sigma(70) region 4, interactions of CAP and RNAP with promoter DNA, and phased-DNA-bend-dependent partial wrapping of DNA around the complex. The structure also reveals the positions and shapes of species-specific domains within the RNAP beta', beta, and sigma(70) subunits.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Rutgers University, 610 Taylor Road, Piscataway, NJ 08854, USA.